
NADH dehydrogenase
Encyclopedia
NADH dehydrogenase (also referred to as NADH:ubiquinone reductase or Complex I) is an enzyme
located in the inner mitochondrial membrane that catalyzes the transfer of electron
s from NADH to coenzyme Q
(CoQ). It is the "entry enzyme" of oxidative phosphorylation
in the mitochondria.
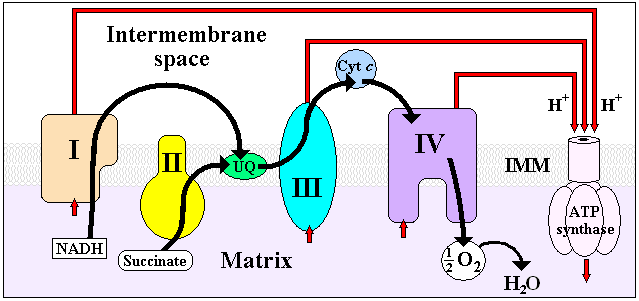
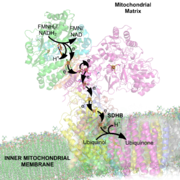 NADH Dehydrogenase is the first enzyme (Complex I) of the mitochondrial electron transport chain
NADH Dehydrogenase is the first enzyme (Complex I) of the mitochondrial electron transport chain
. There are three energy-transducing enzymes in the electron transport chain - NADH dehydrogenase (Complex I), Coenzyme Q – cytochrome c reductase
(Complex III), and cytochrome c oxidase
(Complex IV). NADH dehydrogenase is the largest and most complicated enzyme of the electron transport chain.
The reaction of NADH dehydrogenase is:
In this process, the complex translocates four protons
across the inner membrane per molecule of oxidized NADH, helping to build the electrochemical potential used to produce ATP
.
The reaction can be reversed - referred to as aerobic succinate-supported NAD+ reduction - in the presence of a high membrane potential, but the exact catalytic mechanism remains unknown.
Complex I may have a role in triggering apoptosis
. In fact, there has been shown to be a correlation between mitochondrial activities and programmed cell death
(PCD) during somatic embryo development.
(FMN) prosthetic group of complex I, creating FMNH2. The electron acceptor - the isoalloxazine ring - of FMN is identical to that of FAD
. The electrons are then transferred through the second prosthetic group of NADH dehydrogenase via a series of iron-sulfur (Fe-S) clusters, and finally to coenzyme Q (ubiquinone). This electron flow causes four hydrogen ions to be pumped out of the mitochondrial matrix. Ubiquinone (CoQ) accepts two electrons to be reduced to ubiquionol (CoQH2).
s, the enzyme contains 45 separate polypeptide chains. Of particular functional importance are the flavin prosthetic group (FMN) and eight iron-sulfur cluster
s (FeS). Of the 45 subunits, seven are encoded by the mitochondrial genome.
The structure is an "L" shape with a long membrane domain (with around 60 trans-membrane helices) and a hydrophilic peripheral domain, which includes all the known redox centres and the NADH binding site. Whereas the structure of the eukaryotic complex is not well characterised, the peripheral/hydrophilic domain of the complex from a bacterium (Thermus thermophilus) has been crystallised .
A recent study by Roessler et al. (2010) used electron paramagnetic resonance
(EPR) spectra and double electron-electron resonance (DEER) to determine the path of electron transfer through the iron-sulfur complexes, which are located in the hydrophilic domain. Seven of these clusters form a chain from the flavin to the quinone binding sites; the eighth cluster is located on the other side of the flavin, and its function is unknown. The EPR and DEER results suggest an alternating or “roller-coaster” potential energy profile for the electron transfer between the active sites and along the iron-sulfur clusters, which can optimize the rate of electron travel and allow efficient energy conversion in Complex I.
A simulational study by Hayashi and Stuchebrukhov further identified the electron tunneling pathways in atomic resolution based on the tunneling current theory. The distinct pathways between neighboring Fe/S clusters primarily consist of two cysteine ligands and one additional key residue, which was supported by sensitivity of simulated electron transfer rates to their mutations and their conservation among various complex I homologues from simple bacteria to human beings. This result shows that the crucial part of complex I developed for optimal efficiency with specific key residues during early stages of the biological evolution and has been conserved since then. Internal water between protein subunits was identified as an essential mediator enhancing the overall electron transfer rate to achieve physiologically significant value.
(commonly used as an organic pesticide). Rotenone and rotenoids are isoflavonoids occurring in several genera of tropical plants such as Antonia (Loganiaceae
), Derris
and Lonchocarpus
(Faboideae
, Fabaceae
). There have been reports of Indians using rotenone-containing plants to fish - due to its ichthyotoxic effect - as early as the 17th century. Rotenone binds to the ubiquinone binding site of Complex I as well as piericidin A
, another potent inhibitor with a close structural homologue to ubiquinone.
Despite more than 50 years of study of NADH dehydrogenase, no inhibitors blocking the electron flow inside the enzyme have been found. Hydrophobic inhibitors like rotenone or piericidin most likely disrupt the electron transfer between the terminal FeS cluster N2 and ubiquinone. It has been shown that long-term systemic inhibition of Complex I by rotenone can induce selective degeneration of dopaminergic neurons.
NADH dehydrogenase is also blocked by adenosine diphosphate ribose
- a reversible competitive inhibitor of NADH oxidation by binding to the enzyme at the nucleotide binding site. Both hydrophylic NADH and hydrophobic ubiquinone analogs act at the beginning and the end of the internal electron-transport pathway, respectively.
The acetogenin family are the most potent Complex I inhibitors. They have been shown to crosslink to the ND2 subunit, which suggests that ND2 is essential for quinone-binding. Interestingly, Rolliniastatin-2, an acetogenin, is the first Complex I inhibitor found that does not share the same binding site as rotenone.
The high activation energy
(270 kJ/mol) of the deactivation process indicates the occurrence of major conformational changes in the organisation of the Complex I. However, until now, the only conformational difference observed between these two forms is the number of cysteine
residues exposed at the surface of the enzyme. Treatment of the D-form of complex I with the sulfhydryl reagents N-Ethylmaleimide
or DTNB
irreversibly blocks critical cysteine residue(s), abolishing the ability of the enzyme to respond to activation, thus inactivating it irreversibly. The A-form of complex I is insensitive to sulfhydryl reagents.
It was found that these conformational changes may have a very important physiological significance. The de-active, but not the active form of Complex I was susceptible to inhibition by nitrosothiols and peroxynitrite
. It is likely that transition from the active to the deactive form of complex I takes place during pathological conditions when the turnover of the enzyme is limited at physiological temperatures, such as during hypoxia
, or when the tissue nitric oxide
:oxygen ratio increases (i.e. metabolic hypoxia).
. Complex I can produce superoxide (as well as hydrogen peroxide), through at least two different pathways. During forward electron transfer, only very small amounts of superoxide are produced (probably less than 0.1% of the overall electron flow).
During reverse electron transfer, Complex I might be the most important site of superoxide production within mitochondria, with up to 5% of electrons being diverted to superoxide formation. Reverse electron transfer, the process by which electrons from the reduced ubiquinol pool (supplied by succinate dehydrogenase, glycerol-3-phosphate dehydrogenase
, or dihydro-oorotate dehydrogenase in mammalian mitochondria) pass through Complex I to reduce NAD+ to NADH, driven by the inner mitochondrial membrane potential electric potential. Although it is not precisely known under what pathological conditions reverse-electron transfer would occur in vivo, in vitro experiments indicate that it can be a very potent source of superoxide when succinate concentrations are high and oxaloacetate or malate
concentrations are low.
Superoxide is a reactive oxygen species that contributes to cellular oxidative stress and is linked to neuromuscular diseases and aging. NADH dehdyrogenase produces superoxide by transferring one electron from FMNH2 to oxygen (O2). The radical flavin leftover is unstable, and transfers the remaining electron to the iron-sulfur centers. Interestingly, it is the ratio of NADH to NAD+ that determines the rate of superoxide formation.
. There is some evidence that Complex I defects may play a role in the etiology of Parkinson's disease
, perhaps because of reactive oxygen species (Complex I can, like Complex III, leak electrons to oxygen, forming highly toxic superoxide
).
Although the exact etiology of Parkinson’s disease is unclear, it is likely that mitochondrial dysfunction, along with proteasome inhibition and environmental toxins, may play a large role. In fact, the inhibition of Complex I has been shown to cause the production of peroxides and a decrease in proteasome activity, which may lead to Parkinson’s disease. Additionally, Esteves et al. (2010) found that cell lines with Parkinson’s disease show increased proton leakage in Complex I, which causes decreased maximum respiratory capacity.
Recent studies have examined other roles of NADH dehydrogenase activity in the brain. Andreazza et al. (2010) found that the level of Complex I activity was significantly decreased in patients with bipolar disorder, but not in patients with depression or schizophrenia. They found that patients with bipolar disorder showed increased protein oxidation and nitration in their prefrontal cortex. These results suggest that future studies should target Complex I for potential therapeutic studies for bipolar disorder. Similarly, Moran et al. (2010) found that patients with severe Complex I deficiency showed decreased oxygen consumption rates and slower growth rates. However, they found that mutations in different genes in Complex I lead to different phenotypes, thereby explaining the variations of pathophysiological manifestations of Complex I deficiency.
Exposure to pesticides can also inhibit Complex I and cause disease symptoms. For example, chronic exposure to low levels of dichlorvos, an organophosphate used as a pesticide, has been shown to cause liver dysfunction. This occurs because dichlorvos alters Complex I and II activity levels, which leads to decreased mitochondrial electron transfer activities and decreased ATP synthesis.
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
located in the inner mitochondrial membrane that catalyzes the transfer of electron
Electron
The electron is a subatomic particle with a negative elementary electric charge. It has no known components or substructure; in other words, it is generally thought to be an elementary particle. An electron has a mass that is approximately 1/1836 that of the proton...
s from NADH to coenzyme Q
Coenzyme Q
Coenzyme Q10, also known as ubiquinone, ubidecarenone, coenzyme Q, and abbreviated at times to CoQ10 , CoQ, Q10, or Q, is a 1,4-benzoquinone, where Q refers to the quinone chemical group, and 10 refers to the number of isoprenyl chemical subunits in its tail.This oil-soluble, vitamin-like substance...
(CoQ). It is the "entry enzyme" of oxidative phosphorylation
Oxidative phosphorylation
Oxidative phosphorylation is a metabolic pathway that uses energy released by the oxidation of nutrients to produce adenosine triphosphate . Although the many forms of life on earth use a range of different nutrients, almost all aerobic organisms carry out oxidative phosphorylation to produce ATP,...
in the mitochondria.
Function



Electron transport chain
An electron transport chain couples electron transfer between an electron donor and an electron acceptor with the transfer of H+ ions across a membrane. The resulting electrochemical proton gradient is used to generate chemical energy in the form of adenosine triphosphate...
. There are three energy-transducing enzymes in the electron transport chain - NADH dehydrogenase (Complex I), Coenzyme Q – cytochrome c reductase
Coenzyme Q – cytochrome c reductase
In enzymology, a ubiquinol—cytochrome-c reductase is an enzyme that catalyzes the chemical reactionThus, the two substrates of this enzyme are dihydroquinone and ferri- cytochrome c, whereas its 3 products are quinone , ferro- cytochrome c, and H+.This enzyme belongs to the family of...
(Complex III), and cytochrome c oxidase
Cytochrome c oxidase
The enzyme cytochrome c oxidase or Complex IV is a large transmembrane protein complex found in bacteria and the mitochondrion.It is the last enzyme in the respiratory electron transport chain of mitochondria located in the mitochondrial membrane...
(Complex IV). NADH dehydrogenase is the largest and most complicated enzyme of the electron transport chain.
The reaction of NADH dehydrogenase is:
- NADH + H+ + CoQ + 4H+in → NAD+ + CoQH2 + 4H+out
In this process, the complex translocates four protons
Proton pump
A proton pump is an integral membrane protein that is capable of moving protons across a cell membrane, mitochondrion, or other organelle. Mechanisms are based on conformational changes of the protein structure or on the Q cycle.-Function:...
across the inner membrane per molecule of oxidized NADH, helping to build the electrochemical potential used to produce ATP
Adenosine triphosphate
Adenosine-5'-triphosphate is a multifunctional nucleoside triphosphate used in cells as a coenzyme. It is often called the "molecular unit of currency" of intracellular energy transfer. ATP transports chemical energy within cells for metabolism...
.
The reaction can be reversed - referred to as aerobic succinate-supported NAD+ reduction - in the presence of a high membrane potential, but the exact catalytic mechanism remains unknown.
Complex I may have a role in triggering apoptosis
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
. In fact, there has been shown to be a correlation between mitochondrial activities and programmed cell death
Programmed cell death
Programmed cell-death is death of a cell in any form, mediated by an intracellular program. PCD is carried out in a regulated process which generally confers advantage during an organism's life-cycle...
(PCD) during somatic embryo development.
Mechanism
All redox reactions take place in the extramembranous portion of NADH dehydrogenase. NADH initially binds to NADH dehydrogenase, and transfers two electrons to the flavin mononucleotideFlavin mononucleotide
Flavin mononucleotide , or riboflavin-5′-phosphate, is a biomolecule produced from riboflavin by the enzyme riboflavin kinase and functions as prosthetic group of various oxidoreductases including NADH dehydrogenase as well as cofactor in biological blue-light photo receptors...
(FMN) prosthetic group of complex I, creating FMNH2. The electron acceptor - the isoalloxazine ring - of FMN is identical to that of FAD
FAD
In biochemistry, flavin adenine dinucleotide is a redox cofactor involved in several important reactions in metabolism. FAD can exist in two different redox states, which it converts between by accepting or donating electrons. The molecule consists of a riboflavin moiety bound to the phosphate...
. The electrons are then transferred through the second prosthetic group of NADH dehydrogenase via a series of iron-sulfur (Fe-S) clusters, and finally to coenzyme Q (ubiquinone). This electron flow causes four hydrogen ions to be pumped out of the mitochondrial matrix. Ubiquinone (CoQ) accepts two electrons to be reduced to ubiquionol (CoQH2).
Composition and structure
NADH Dehydrogenase is the largest of the respiratory complexes. In mammalMammal
Mammals are members of a class of air-breathing vertebrate animals characterised by the possession of endothermy, hair, three middle ear bones, and mammary glands functional in mothers with young...
s, the enzyme contains 45 separate polypeptide chains. Of particular functional importance are the flavin prosthetic group (FMN) and eight iron-sulfur cluster
Iron-sulfur cluster
For biological Fe-S clusters, see iron-sulfur proteins.Iron-sulfur clusters are ensembles of iron and sulfide centres. Fe-S clusters are most often discussed in the context of the biological role for iron-sulfur proteins. Many Fe-S clusters are known in the area of organometallic chemistry and as...
s (FeS). Of the 45 subunits, seven are encoded by the mitochondrial genome.
The structure is an "L" shape with a long membrane domain (with around 60 trans-membrane helices) and a hydrophilic peripheral domain, which includes all the known redox centres and the NADH binding site. Whereas the structure of the eukaryotic complex is not well characterised, the peripheral/hydrophilic domain of the complex from a bacterium (Thermus thermophilus) has been crystallised .
A recent study by Roessler et al. (2010) used electron paramagnetic resonance
Electron paramagnetic resonance
Electron paramagnetic resonance or electron spin resonance spectroscopyis a technique for studying chemical species that have one or more unpaired electrons, such as organic and inorganic free radicals or inorganic complexes possessing a transition metal ion...
(EPR) spectra and double electron-electron resonance (DEER) to determine the path of electron transfer through the iron-sulfur complexes, which are located in the hydrophilic domain. Seven of these clusters form a chain from the flavin to the quinone binding sites; the eighth cluster is located on the other side of the flavin, and its function is unknown. The EPR and DEER results suggest an alternating or “roller-coaster” potential energy profile for the electron transfer between the active sites and along the iron-sulfur clusters, which can optimize the rate of electron travel and allow efficient energy conversion in Complex I.
A simulational study by Hayashi and Stuchebrukhov further identified the electron tunneling pathways in atomic resolution based on the tunneling current theory. The distinct pathways between neighboring Fe/S clusters primarily consist of two cysteine ligands and one additional key residue, which was supported by sensitivity of simulated electron transfer rates to their mutations and their conservation among various complex I homologues from simple bacteria to human beings. This result shows that the crucial part of complex I developed for optimal efficiency with specific key residues during early stages of the biological evolution and has been conserved since then. Internal water between protein subunits was identified as an essential mediator enhancing the overall electron transfer rate to achieve physiologically significant value.
Inhibitors
The best-known inhibitor of Complex I is rotenoneRotenone
Rotenone is an odorless chemical that is used as a broad-spectrum insecticide, piscicide, and pesticide. It occurs naturally in the roots and stems of several plants such as the jicama vine plant...
(commonly used as an organic pesticide). Rotenone and rotenoids are isoflavonoids occurring in several genera of tropical plants such as Antonia (Loganiaceae
Loganiaceae
Loganiaceae are a family of flowering plants classified in order Gentianales. The family includes 13 genera, distributed around the world's tropics.Earlier treatments of the family have included up to 29 genera...
), Derris
Derris
Derris is a climbing leguminous plant of Southeast Asia and the southwest Pacific islands, including New Guinea. Its roots contain rotenone, a strong insecticide and fish poison....
and Lonchocarpus
Lonchocarpus
Lonchocarpus is a plant genus in the legume family . The species are called lancepods due to their fruit resembling an ornate lance tip or a few beads on a string....
(Faboideae
Faboideae
Faboideae is a subfamily of the flowering plant family Fabaceae or Leguminosae. One acceptable alternative name for the subfamily is Papilionoideae....
, Fabaceae
Fabaceae
The Fabaceae or Leguminosae, commonly known as the legume, pea, or bean family, is a large and economically important family of flowering plants. The group is the third largest land plant family, behind only the Orchidaceae and Asteraceae, with 730 genera and over 19,400 species...
). There have been reports of Indians using rotenone-containing plants to fish - due to its ichthyotoxic effect - as early as the 17th century. Rotenone binds to the ubiquinone binding site of Complex I as well as piericidin A
Piericidin A
Piericidin A is an inhibitor of NADH dehydrogenase.Piericidin A Inhibits electron transfer; it competes with QB for binding site in Photosystem II....
, another potent inhibitor with a close structural homologue to ubiquinone.
Despite more than 50 years of study of NADH dehydrogenase, no inhibitors blocking the electron flow inside the enzyme have been found. Hydrophobic inhibitors like rotenone or piericidin most likely disrupt the electron transfer between the terminal FeS cluster N2 and ubiquinone. It has been shown that long-term systemic inhibition of Complex I by rotenone can induce selective degeneration of dopaminergic neurons.
NADH dehydrogenase is also blocked by adenosine diphosphate ribose
Adenosine diphosphate ribose
Adenosine diphosphate ribose is a molecule formed into chains by the enzyme poly ADP ribose polymerase. It binds to and activates the TRPM2 ion channel....
- a reversible competitive inhibitor of NADH oxidation by binding to the enzyme at the nucleotide binding site. Both hydrophylic NADH and hydrophobic ubiquinone analogs act at the beginning and the end of the internal electron-transport pathway, respectively.
The acetogenin family are the most potent Complex I inhibitors. They have been shown to crosslink to the ND2 subunit, which suggests that ND2 is essential for quinone-binding. Interestingly, Rolliniastatin-2, an acetogenin, is the first Complex I inhibitor found that does not share the same binding site as rotenone.
Active/de-active transition
The catalytic properties of eukaryotic Complex I are not simple. Two catalytically and structurally distinct forms exist in any given preparation of the enzyme: one is the fully competent, so-called “active” A-form and the other is the catalytically silent, dormant, “de-activated”, D-form. After exposure of idle enzyme to elevated, but physiological temperatures (>30°C) in the absence of substrate, the enzyme converts to the D-form. This form is catalytically incompetent but can be activated by the slow reaction (k~4 min−1) of NADH oxidation with subsequent ubiquinone reduction. After one or several turnovers the enzyme becomes active and can catalyse physiological NADH:ubiquinone reaction at a much higher rate (k~104 min−1). In the presence of divalent cations (Mg2+, Ca2+), or at alkaline pH the activation takes much longer.The high activation energy
Activation energy
In chemistry, activation energy is a term introduced in 1889 by the Swedish scientist Svante Arrhenius that is defined as the energy that must be overcome in order for a chemical reaction to occur. Activation energy may also be defined as the minimum energy required to start a chemical reaction...
(270 kJ/mol) of the deactivation process indicates the occurrence of major conformational changes in the organisation of the Complex I. However, until now, the only conformational difference observed between these two forms is the number of cysteine
Cysteine
Cysteine is an α-amino acid with the chemical formula HO2CCHCH2SH. It is a non-essential amino acid, which means that it is biosynthesized in humans. Its codons are UGU and UGC. The side chain on cysteine is thiol, which is polar and thus cysteine is usually classified as a hydrophilic amino acid...
residues exposed at the surface of the enzyme. Treatment of the D-form of complex I with the sulfhydryl reagents N-Ethylmaleimide
N-Ethylmaleimide
N-Ethylmaleimide is an organic compound that is derived from maleic acid. It contains the imide functional group, but more importantly it is an alkene that is reactive toward thiols and is commonly used to modify cysteine residues in proteins and peptides.-Organic chemistry:In the jargon of...
or DTNB
DTNB
Beta dystrobrevin also known as DTNB is a protein which in humans is encoded by the DTNB gene.- Function :This gene encodes dystrobrevin beta, a component of the dystrophin-associated protein complex . The DPC consists of dystrophin and several integral and peripheral membrane proteins, including...
irreversibly blocks critical cysteine residue(s), abolishing the ability of the enzyme to respond to activation, thus inactivating it irreversibly. The A-form of complex I is insensitive to sulfhydryl reagents.
It was found that these conformational changes may have a very important physiological significance. The de-active, but not the active form of Complex I was susceptible to inhibition by nitrosothiols and peroxynitrite
Peroxynitrite
Peroxynitrite is the anion with the formula ONOO−. It is an unstable structural isomer of nitrate, NO3−, which has the same formula but a different structure. Although peroxynitrous acid is highly reactive, its conjugate base peroxynitrite is stable in basic solution...
. It is likely that transition from the active to the deactive form of complex I takes place during pathological conditions when the turnover of the enzyme is limited at physiological temperatures, such as during hypoxia
Hypoxia (medical)
Hypoxia, or hypoxiation, is a pathological condition in which the body as a whole or a region of the body is deprived of adequate oxygen supply. Variations in arterial oxygen concentrations can be part of the normal physiology, for example, during strenuous physical exercise...
, or when the tissue nitric oxide
Nitric oxide
Nitric oxide, also known as nitrogen monoxide, is a diatomic molecule with chemical formula NO. It is a free radical and is an important intermediate in the chemical industry...
:oxygen ratio increases (i.e. metabolic hypoxia).
Production of superoxide
Recent investigations suggest that Complex I is a potent source of reactive oxygen speciesReactive oxygen species
Reactive oxygen species are chemically reactive molecules containing oxygen. Examples include oxygen ions and peroxides. Reactive oxygen species are highly reactive due to the presence of unpaired valence shell electrons....
. Complex I can produce superoxide (as well as hydrogen peroxide), through at least two different pathways. During forward electron transfer, only very small amounts of superoxide are produced (probably less than 0.1% of the overall electron flow).
During reverse electron transfer, Complex I might be the most important site of superoxide production within mitochondria, with up to 5% of electrons being diverted to superoxide formation. Reverse electron transfer, the process by which electrons from the reduced ubiquinol pool (supplied by succinate dehydrogenase, glycerol-3-phosphate dehydrogenase
Glycerol-3-phosphate dehydrogenase
Glycerol-3-phosphate dehydrogenase is an enzyme that catalyzes the reversible redox conversion of dihydroxyacetone phosphate to sn-glycerol 3-phosphate....
, or dihydro-oorotate dehydrogenase in mammalian mitochondria) pass through Complex I to reduce NAD+ to NADH, driven by the inner mitochondrial membrane potential electric potential. Although it is not precisely known under what pathological conditions reverse-electron transfer would occur in vivo, in vitro experiments indicate that it can be a very potent source of superoxide when succinate concentrations are high and oxaloacetate or malate
Malate
Malate is the ionized form of malic acid. It is an important chemical compound in biochemistry. In the C4 carbon fixation process, malate is a source of CO2 in the Calvin cycle....
concentrations are low.
Superoxide is a reactive oxygen species that contributes to cellular oxidative stress and is linked to neuromuscular diseases and aging. NADH dehdyrogenase produces superoxide by transferring one electron from FMNH2 to oxygen (O2). The radical flavin leftover is unstable, and transfers the remaining electron to the iron-sulfur centers. Interestingly, it is the ratio of NADH to NAD+ that determines the rate of superoxide formation.
Pathology
Mutations in the subunits of Complex I can cause mitochondrial diseases, including Leigh syndrome. Point mutations in various Complex I subunits derived from mitochondrial DNA (mtDNA) can also result in Leber's Hereditary Optic NeuropathyLeber's hereditary optic neuropathy
Leber’s hereditary optic neuropathy or Leber optic atrophy is a mitochondrially inherited degeneration of retinal ganglion cells and their axons that leads to an acute or subacute loss of central vision; this affects predominantly young adult males...
. There is some evidence that Complex I defects may play a role in the etiology of Parkinson's disease
Parkinson's disease
Parkinson's disease is a degenerative disorder of the central nervous system...
, perhaps because of reactive oxygen species (Complex I can, like Complex III, leak electrons to oxygen, forming highly toxic superoxide
Superoxide
A superoxide, also known by the obsolete name hyperoxide, is a compound that possesses the superoxide anion with the chemical formula O2−. The systematic name of the anion is dioxide. It is important as the product of the one-electron reduction of dioxygen O2, which occurs widely in nature...
).
Although the exact etiology of Parkinson’s disease is unclear, it is likely that mitochondrial dysfunction, along with proteasome inhibition and environmental toxins, may play a large role. In fact, the inhibition of Complex I has been shown to cause the production of peroxides and a decrease in proteasome activity, which may lead to Parkinson’s disease. Additionally, Esteves et al. (2010) found that cell lines with Parkinson’s disease show increased proton leakage in Complex I, which causes decreased maximum respiratory capacity.
Recent studies have examined other roles of NADH dehydrogenase activity in the brain. Andreazza et al. (2010) found that the level of Complex I activity was significantly decreased in patients with bipolar disorder, but not in patients with depression or schizophrenia. They found that patients with bipolar disorder showed increased protein oxidation and nitration in their prefrontal cortex. These results suggest that future studies should target Complex I for potential therapeutic studies for bipolar disorder. Similarly, Moran et al. (2010) found that patients with severe Complex I deficiency showed decreased oxygen consumption rates and slower growth rates. However, they found that mutations in different genes in Complex I lead to different phenotypes, thereby explaining the variations of pathophysiological manifestations of Complex I deficiency.
Exposure to pesticides can also inhibit Complex I and cause disease symptoms. For example, chronic exposure to low levels of dichlorvos, an organophosphate used as a pesticide, has been shown to cause liver dysfunction. This occurs because dichlorvos alters Complex I and II activity levels, which leads to decreased mitochondrial electron transfer activities and decreased ATP synthesis.
Genes
The following is a list of humans genes that encode components of the NADH dehydrogenase (ubiquinone) complex:- NADH dehydrogenase (ubiquinone) 1 alpha subcomplex
- NDUFA1NDUFA1NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 is a protein that in humans is encoded by the NDUFA1 gene.- Function :The human NDUFA1 gene codes for an essential component of complex I of the respiratory chain, which transfers electrons from NADH to ubiquinone...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa - NDUFA2NDUFA2NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 is an enzyme that in humans is encoded by the NDUFA2 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa - NDUFA3 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
- NDUFA4 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
- NDUFA4L – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like
- NDUFA4L2 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
- NDUFA5NDUFA5NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 is an enzyme that in humans is encoded by the NDUFA5 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa - NDUFA6NDUFA6NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 is an enzyme that in humans is encoded by the NDUFA6 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa - NDUFA7 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa
- NDUFA8NDUFA8NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 is an enzyme that in humans is encoded by the NDUFA8 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa - NDUFA9NDUFA9NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial is an enzyme that in humans is encoded by the NDUFA9 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa - NDUFA10NDUFA10NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial is an enzyme that in humans is encoded by the NDUFA10 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa - NDUFA11 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa
- NDUFA12NDUFA12NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 is an enzyme that in humans is encoded by the NDUFA12 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 - NDUFA13NDUFA13NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 is an enzyme that in humans is encoded by the NDUFA13 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 - NDUFAB1 – NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa
- NDUFAF1NDUFAF1Complex I intermediate-associated protein 30, mitochondrial is a protein that in humans is encoded by the NDUFAF1 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 - NDUFAF2 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 2
- NDUFAF3 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
- NDUFAF4 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
- NDUFA1
- NADH dehydrogenase (ubiquinone) 1 beta subcomplex
- NDUFB1NDUFB1NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 is an enzyme that in humans is encoded by the NDUFB1 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa - NDUFB2NDUFB2NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2, mitochondrial is an enzyme that in humans is encoded by the NDUFB2 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa - NDUFB3 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa
- NDUFB4 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa
- NDUFB5 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa
- NDUFB6NDUFB6NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6 also known as complex I-B17 is a protein that in humans is encoded by the NDUFB6 gene.- Function :...
– NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa - NDUFB7NDUFB7NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 is an enzyme that in humans is encoded by the NDUFB7 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa - NDUFB8NDUFB8NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial is an enzyme that in humans is encoded by the NDUFB8 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa - NDUFB9NDUFB9NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 is an enzyme that in humans is encoded by the NDUFB9 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa - NDUFB10NDUFB10NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 is an enzyme that in humans is encoded by the NDUFB10 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa - NDUFB11NDUFB11-Further reading:...
– NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa
- NDUFB1
- NADH dehydrogenase (ubiquinone) 1, subcomplex unknown
- NDUFC1NDUFC1NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial is an enzyme that in humans is encoded by the NDUFC1 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa - NDUFC2NDUFC2NADH dehydrogenase [ubiquinone] 1 subunit C2 is an enzyme that in humans is encoded by the NDUFC2 gene.The NDUF2 gene encodes one of the subunits of complex I, the first and largest complex of the mitochondrial respiratory chain.-Further reading:...
– NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 2, 14.5kDa
- NDUFC1
- NADH dehydrogenase (ubiquinone) Fe-S protein
- NDUFS1NDUFS1NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial is an enzyme that in humans is encoded by the NDUFS1 gene.- Function :The protein encoded by this gene belongs to the complex I 75 kDa subunit family. Mammalian complex I is composed of 45 different subunits. It locates at the...
– NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) - NDUFS2NDUFS2NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial also known as NADH-ubiquinone oxidoreductase 49 kDa subunit is an enzyme that in humans is encoded by the NDUFS2 gene.- Function :...
– NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase) - NDUFS3NDUFS3NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial is an enzyme that in humans is encoded by the NDUFS3 gene.- Function :...
– NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) - NDUFS4NDUFS4NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial also known as NADH-ubiquinone oxidoreductase 18 kDa subunit is an enzyme that in humans is encoded by the NDUFS4 gene.- Function :...
– NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase) - NDUFS5NDUFS5NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 is an enzyme that in humans is encoded by the NDUFS5 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) - NDUFS6NDUFS6NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial is an enzyme that in humans is encoded by the NDUFS6 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) - NDUFS7NDUFS7NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial is an enzyme that in humans is encoded by the NDUFS7 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) - NDUFS8NDUFS8NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial also known as NADH-ubiquinone oxidoreductase 23 kDa subunit is an enzyme that in humans is encoded by the NDUFS8 gene.- Function :...
– NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase)
- NDUFS1
- NADH dehydrogenase (ubiquinone) flavoprotein 1
- NDUFV1NDUFV1NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial is an enzyme that in humans is encoded by the NDUFV1 gene.The NDUFV1 gene encodes the 51-kD subunit of complex I of the mitochondrial respiratory chain....
– NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa - NDUFV2NDUFV2NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial is an enzyme that in humans is encoded by the NDUFV2 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa - NDUFV3NDUFV3NADH dehydrogenase [ubiquinone] flavoprotein 3, mitochondrial is an enzyme that in humans is encoded by the NDUFV3 gene.-Further reading:...
– NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa
- NDUFV1
- mitochondrially encoded NADH dehydrogenase subunit
- MT-ND1MT-ND1MT-ND1 is a mitochondrial gene. It is associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes.-External links:*...
- mitochondrially encoded NADH dehydrogenase subunit 1 - MT-ND2 - mitochondrially encoded NADH dehydrogenase subunit 2
- MT-ND3 - mitochondrially encoded NADH dehydrogenase subunit 3
- MT-ND4 - mitochondrially encoded NADH dehydrogenase subunit 4
- MT-ND4L - mitochondrially encoded NADH dehydrogenase subunit 4L
- MT-ND5MT-ND5MT-ND5 is a mitochondrial gene. It is associated with MELAS or mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes.-External links:*...
- mitochondrially encoded NADH dehydrogenase subunit 5 - MT-ND6 - mitochondrially encoded NADH dehydrogenase subunit 6
- MT-ND1
External links
- Interactive Molecular model of NADH dehydrogenase (Requires MDL Chime)
- Complex I home page at The Scripps Research InstituteThe Scripps Research InstituteThe Scripps Research Institute is an American medical research facility that focuses on research in the basic biomedical sciences. Headquartered in La Jolla, California, with a sister facility in Jupiter, Florida, the institute is home to 3,000 scientists, technicians, graduate students, and...

