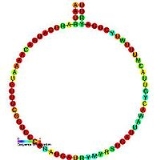
SnoRNA
Encyclopedia
Small nucleolar RNAs are a class of small RNA
molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNA
s, transfer RNA
s and small nuclear RNA
s. There are two main classes of snoRNA, the C/D box snoRNAs which are associated with methylation
, and the H/ACA box snoRNAs which are associated with pseudouridylation.
snoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNA
s that direct RNA editing in trypanosomes.
, nascent rRNA molecules (termed pre-rRNA) are required to undergo a series of processing steps in order to generate the mature rRNA molecule. Prior to cleavage by exo- and endonucleases the pre-rRNA undergoes a complex pattern of nucleoside modifications. These include methylations and pseudouridylations, guided by snoRNAs.
Each snoRNA molecule acts as a guide for only one (or two) individual modifications in a target RNA. In order to carry out modification, each snoRNA associates with at least four protein molecules in an RNA/protein complex referred to as a small nucleolar ribonucleoprotein (snoRNP). The proteins associated with each RNA depend on the type of snoRNA molecule (see snoRNA guide families below). The snoRNA molecule contains an antisense element (a stretch of 10-20 nucleotides) which are base complementary to the sequence surrounding the base (nucleotide
) targeted for modification in the pre-RNA molecule. This enables the snoRNP to recognise and bind to the target RNA. Once the snoRNP has bound to the target site the associated proteins are in the correct physical location to catalyse the chemical modification of the target base.
SnoRNAs are classified under small nuclear RNA in MeSH
.
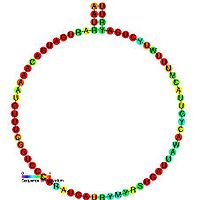 C/D box snoRNAs contain two short conserved sequence motifs, C (UGAUGA) and D (CUGA) located near the 5' and 3' ends of the snoRNA respectively. Short regions (~ 5 nucleotides) located upstream
C/D box snoRNAs contain two short conserved sequence motifs, C (UGAUGA) and D (CUGA) located near the 5' and 3' ends of the snoRNA respectively. Short regions (~ 5 nucleotides) located upstream
of the C box and downstream
of the D box are usually base complementary and form a stem-box structure which brings the C and D box motifs into close proximity. This stem-box structure has been shown to be essential for correct snoRNA synthesis and nucleolar localization. Many C/D box snoRNA also contain an additional less well conserved copy of the C and D motifs (referred to as C' and D') located in the central portion of the snoRNA molecule. A conserved region of 10-21 nucleotides upstream of the D box is complementary to the methylation site of the target RNA and enables the snoRNA to form an RNA duplex with the RNA. The nucleotide to be modified in the target RNA is usually located at the 5th position upstream from the D box (or D' box). C/D box snoRNAs associate with four evolutionary conserved and essential proteins ( Fibrillarin
(Nop1p), Nop56p, Nop58p, and Snu13 (in eukaryotes; its archaeal homolog is L7Ae)) which make up the core C/D box snoRNP.
There exists a eukaryotic C/D box snoRNA (snoRNA U3) that has not been shown to guide 2'-O-methylation.
Instead, it functions in rRNA processing by directing pre-rRNA cleavage.
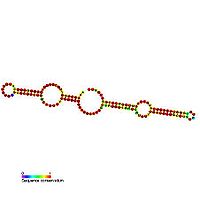 H/ACA box snoRNAs have a common secondary structure
H/ACA box snoRNAs have a common secondary structure
consisting of a two hairpins and two single stranded regions termed a hairpin-hinge-hairpin-tail structure. H/ACA snoRNAs also contain conserved sequence motifs known as H box (consensus ANANNA) and the ACA box (ACA). Both motifs are usually located in the single stranded regions of the secondary structure. The H motif is located in the hinge and the ACA motif is located in the tail region, 3 nucleotides from the 3' end of the sequence. The hairpin regions contain internal bulges known as recognition loops in which the antisense guide sequences (bases complementary to the target sequence) are located. This recognition sequence is bipartite (constructed from the two different arms of the loop region) and forms complex pseudo-knots
with the target RNA. H/ACA box snoRNAs associate with four evolutionary conserved and essential proteins ( dyskerin (Cbf5p), Gar1p, Nhp2p and Nop10p) which make up the core of the H/ACA box snoRNP. However, in lower eukaryotic cells such as trypanosomes, similar RNAs exist in the form of single hairpin structure and an AGA box instead of ACA box at the 3' end of the RNA.
The RNA component of human telomerase
(hTERC) contains an H/ACA domain for pre-RNP
formation and nucleolar
localization of the telomerase RNP itself. Importantly, the H/ACA snoRNP has been implicated in the rare genetic disease dyskeratosis congenita
(DKC) due to its affiliation with human telomerase. Mutations in the protein component of the H/ACA snoRNP result in a reduction in physiological TERC levels. This has been strongly correlated with the pathology behind DKC which seems to be primarily a disease of poor telomere
maintenance.
(snRNA) U5. This composite snoRNA contains both C/D and H/ACA box domains and associates with the proteins specific to each class of snoRNA (fibrillaring and Gar1p respectively. More composite snoRNAs have now been characterised.
These composite snoRNAs have been found to accumulate in a subnuclear organelle called the Cajal body
and are referred to as Cajal body specific RNAs. This is in contrast to the majority of C/D box or H/ACA box snoRNAs which localise to the nucleolus. These Cajal body specific RNAs and are proposed to be involved in the modification of RNA polymerase II transcribed spliceosomal RNAs U1, U2, U4, U5 and U12. Not all snoRNAs that have been localised to Cajal bodies are composite C/D and H/ACA box snoRNAs.
regulates the alternative splicing of the serotonin 2C receptor
mRNA via a conserved region of complementarity.
Another C/D box snoRNA, SNORD116
, that resides in the same cluster as SNORD115 has been predicted to have 23 possible targets within protein coding genes using a bioinformatic approach. Of these a large fraction were found to be alternatively spliced, suggesting a role of SNORD116 in the regulation of alternative splicing.
In 15q11q13, five different snoRNAs have been identified (SNORD64
, SNORD107, SNORD108, SNORD109 (two copies), SNORD116
(29 copies) and SNORD115
(48 copies). Loss of the 29 copies of SNORD116 (HBII-85) from this region has been identified as a cause of Prader-Willi syndrome
whereas gain of additional copies of SNORD115 has been linked to autism
.
Region 14q32 contains repeats of two snoRNAs SNORD113
(9 copies) and SNORD114 (31 copies) within the introns of a tissue-specific ncRNA transcript (MEG8
). The 14q32 domain has been shown to share common genomic features with the imprinted 15q11-q13 loci and a possible role for tandem repeats of C/D box snoRNAs in the evolution or mechanism of imprinted loci has been suggested.
s. It has been shown that human ACA45
is a bona fide snoRNA that can be processed into a 21 nucleotides long mature miRNA by the RNAse III family endoribonuclease
dicer
. This snoRNA product has previously been identified as mmu-miR-1839 and was shown to be processed independent of the other miRNA generating endoribonuclease
drosha
. Bioinformatical
analyses have revealed that putatively snoRNA-derived, miRNA-like fragments occur in different organisms.
Recently, it has been found that snoRNAs can have functions not related to rRNA. One such function is the regulation of alternative splicing
of the trans gene transcript, which is done by the snoRNA HBII-52
, which is also known as SNORD115.
Recent discovery also show, the existence of snoRNA, microRNA, piRNA characteristics in a novel non-coding RNA: x-ncRNA and its biological implication in Homo sapiens.
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNA
Ribosomal RNA
Ribosomal ribonucleic acid is the RNA component of the ribosome, the enzyme that is the site of protein synthesis in all living cells. Ribosomal RNA provides a mechanism for decoding mRNA into amino acids and interacts with tRNAs during translation by providing peptidyl transferase activity...
s, transfer RNA
Transfer RNA
Transfer RNA is an adaptor molecule composed of RNA, typically 73 to 93 nucleotides in length, that is used in biology to bridge the three-letter genetic code in messenger RNA with the twenty-letter code of amino acids in proteins. The role of tRNA as an adaptor is best understood by...
s and small nuclear RNA
Small nuclear RNA
Small nuclear ribonucleic acid is a class of small RNA molecules that are found within the nucleus of eukaryotic cells. They are transcribed by RNA polymerase II or RNA polymerase III and are involved in a variety of important processes such as RNA splicing , regulation of transcription factors ...
s. There are two main classes of snoRNA, the C/D box snoRNAs which are associated with methylation
Methylation
In the chemical sciences, methylation denotes the addition of a methyl group to a substrate or the substitution of an atom or group by a methyl group. Methylation is a form of alkylation with, to be specific, a methyl group, rather than a larger carbon chain, replacing a hydrogen atom...
, and the H/ACA box snoRNAs which are associated with pseudouridylation.
snoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNA
Guide RNA
Guide RNAs are the RNAs that guide the insertion or deletion of uridine residues into mitochondrial mRNAs in kinetoplastid protists in a process known as RNA editing.-Overview of gRNA-directed editing:...
s that direct RNA editing in trypanosomes.
snoRNA guided modifications
After transcriptionTranscription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
, nascent rRNA molecules (termed pre-rRNA) are required to undergo a series of processing steps in order to generate the mature rRNA molecule. Prior to cleavage by exo- and endonucleases the pre-rRNA undergoes a complex pattern of nucleoside modifications. These include methylations and pseudouridylations, guided by snoRNAs.
- Methylation is the attachment or substitution of a methyl group onto various substrates. The rRNA of humans contain approximately 115 methyl group modifications. The majority of these are 2'O-ribose-methylation2'-O-methylation2'-O-methylation is a common nucleoside modification of RNA, where a methyl group is added to the 2' hydroxyl group of the ribose moiety of a nucleoside. 2'-O-methylated nucleosides are found in the functionally essential regions of the ribosome and spliceosome. Also, 2'-O-methylation of adenosine...
s ( where the methyl group is attached to the ribose group).
- Pseudouridylation is the conversion (isomerisationIsomerisationIn chemistry isomerisation is the process by which one molecule is transformed into another molecule which has exactly the same atoms, but the atoms are rearranged e.g. A-B-C → B-A-C . In some molecules and under some conditions, isomerisation occurs spontaneously...
) of the nucleoside uridineUridineUridine is a molecule that is formed when uracil is attached to a ribose ring via a β-N1-glycosidic bond.If uracil is attached to a deoxyribose ring, it is known as a deoxyuridine....
to a different isomeric form pseudouridinePseudouridinePseudouridine is the C-glycoside isomer of the nucleoside uridine, and it is the most prevalent of the over one hundred different modified nucleosides found in RNA. Ψ is found in all species and in many classes of RNA except mRNA...
(Ψ). Mature human rRNAs contain approximately 95 Ψ modifications.
Each snoRNA molecule acts as a guide for only one (or two) individual modifications in a target RNA. In order to carry out modification, each snoRNA associates with at least four protein molecules in an RNA/protein complex referred to as a small nucleolar ribonucleoprotein (snoRNP). The proteins associated with each RNA depend on the type of snoRNA molecule (see snoRNA guide families below). The snoRNA molecule contains an antisense element (a stretch of 10-20 nucleotides) which are base complementary to the sequence surrounding the base (nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
) targeted for modification in the pre-RNA molecule. This enables the snoRNP to recognise and bind to the target RNA. Once the snoRNP has bound to the target site the associated proteins are in the correct physical location to catalyse the chemical modification of the target base.
snoRNA guide families
The two different types of rRNA modification (methylation and pseudouridylation) are directed by two different families of snoRNPs. These families of snoRNAs are referred to as antisense C/D box and H/ACA box snoRNAs based on the presence of conserved sequence motifs in the snoRNA. There are exceptions but as a general rule C/D box members guide methylation and H/ACA members guide pseudouridylation. The members of each family may vary in biogenesis, structure and function but each family is classified by the following generalised characteristics. For more detail see review.SnoRNAs are classified under small nuclear RNA in MeSH
Medical Subject Headings
Medical Subject Headings is a comprehensive controlled vocabulary for the purpose of indexing journal articles and books in the life sciences; it can also serve as a thesaurus that facilitates searching...
.
C/D box

Upstream and downstream (DNA)
In molecular biology and genetics, upstream and downstream both refer to a relative position in DNA or RNA. Each strand of DNA or RNA has a 5' end and a 3' end, so named for the carbons on the deoxyribose ring. Relative to the position on the strand, downstream is the region towards the 3' end of...
of the C box and downstream
Upstream and downstream (DNA)
In molecular biology and genetics, upstream and downstream both refer to a relative position in DNA or RNA. Each strand of DNA or RNA has a 5' end and a 3' end, so named for the carbons on the deoxyribose ring. Relative to the position on the strand, downstream is the region towards the 3' end of...
of the D box are usually base complementary and form a stem-box structure which brings the C and D box motifs into close proximity. This stem-box structure has been shown to be essential for correct snoRNA synthesis and nucleolar localization. Many C/D box snoRNA also contain an additional less well conserved copy of the C and D motifs (referred to as C' and D') located in the central portion of the snoRNA molecule. A conserved region of 10-21 nucleotides upstream of the D box is complementary to the methylation site of the target RNA and enables the snoRNA to form an RNA duplex with the RNA. The nucleotide to be modified in the target RNA is usually located at the 5th position upstream from the D box (or D' box). C/D box snoRNAs associate with four evolutionary conserved and essential proteins ( Fibrillarin
Fibrillarin
Fibrillarin is a component of several ribonucleoproteins including a nucleolar small nuclear ribonucleoprotein and one of the two classes of small nucleolar ribonucleoproteins . SnRNAs function in RNA splicing while snoRNPs function in ribosomal RNA processing.Fibrillarin is associated with U3, U8...
(Nop1p), Nop56p, Nop58p, and Snu13 (in eukaryotes; its archaeal homolog is L7Ae)) which make up the core C/D box snoRNP.
There exists a eukaryotic C/D box snoRNA (snoRNA U3) that has not been shown to guide 2'-O-methylation.
Instead, it functions in rRNA processing by directing pre-rRNA cleavage.
H/ACA box

Secondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
consisting of a two hairpins and two single stranded regions termed a hairpin-hinge-hairpin-tail structure. H/ACA snoRNAs also contain conserved sequence motifs known as H box (consensus ANANNA) and the ACA box (ACA). Both motifs are usually located in the single stranded regions of the secondary structure. The H motif is located in the hinge and the ACA motif is located in the tail region, 3 nucleotides from the 3' end of the sequence. The hairpin regions contain internal bulges known as recognition loops in which the antisense guide sequences (bases complementary to the target sequence) are located. This recognition sequence is bipartite (constructed from the two different arms of the loop region) and forms complex pseudo-knots
Pseudoknot
A pseudoknot is a nucleic acid secondary structure containing at least two stem-loop structures in which half of one stem is intercalated between the two halves of another stem. The pseudoknot was first recognized in the turnip yellow mosaic virus in 1982...
with the target RNA. H/ACA box snoRNAs associate with four evolutionary conserved and essential proteins ( dyskerin (Cbf5p), Gar1p, Nhp2p and Nop10p) which make up the core of the H/ACA box snoRNP. However, in lower eukaryotic cells such as trypanosomes, similar RNAs exist in the form of single hairpin structure and an AGA box instead of ACA box at the 3' end of the RNA.
The RNA component of human telomerase
Telomerase
Telomerase is an enzyme that adds DNA sequence repeats to the 3' end of DNA strands in the telomere regions, which are found at the ends of eukaryotic chromosomes. This region of repeated nucleotide called telomeres contains non-coding DNA material and prevents constant loss of important DNA from...
(hTERC) contains an H/ACA domain for pre-RNP
Ribonucleoprotein
Ribonucleoprotein is a nucleoprotein that contains RNA, i.e. it is an association that combines ribonucleic acid and protein together. A few known examples include the ribosome, the enzyme telomerase, vault ribonucleoproteins, and small nuclear RNPs , which are implicated in pre-mRNA splicing and...
formation and nucleolar
Nucleolus
The nucleolus is a non-membrane bound structure composed of proteins and nucleic acids found within the nucleus. Ribosomal RNA is transcribed and assembled within the nucleolus...
localization of the telomerase RNP itself. Importantly, the H/ACA snoRNP has been implicated in the rare genetic disease dyskeratosis congenita
Dyskeratosis congenita
Dyskeratosis congenita , also called Zinsser-Cole-Engman syndrome, is a rare progressive congenital disorder which results in what in some ways resembles premature aging...
(DKC) due to its affiliation with human telomerase. Mutations in the protein component of the H/ACA snoRNP result in a reduction in physiological TERC levels. This has been strongly correlated with the pathology behind DKC which seems to be primarily a disease of poor telomere
Telomere
A telomere is a region of repetitive DNA sequences at the end of a chromosome, which protects the end of the chromosome from deterioration or from fusion with neighboring chromosomes. Its name is derived from the Greek nouns telos "end" and merοs "part"...
maintenance.
Composite H/ACA and C/D box
An unusual guide snoRNA U85 was identified that functions in both 2'-O-ribose methylation and pseudouridylation of small nuclear RNASmall nuclear RNA
Small nuclear ribonucleic acid is a class of small RNA molecules that are found within the nucleus of eukaryotic cells. They are transcribed by RNA polymerase II or RNA polymerase III and are involved in a variety of important processes such as RNA splicing , regulation of transcription factors ...
(snRNA) U5. This composite snoRNA contains both C/D and H/ACA box domains and associates with the proteins specific to each class of snoRNA (fibrillaring and Gar1p respectively. More composite snoRNAs have now been characterised.
These composite snoRNAs have been found to accumulate in a subnuclear organelle called the Cajal body
Cajal body
Cajal bodies are spherical sub-organelles of 0.3-1.0 µm in diameter found in the nucleus of proliferative cells like embryonic cells and tumor cells, or metabolically active cells like neurons. In contrast to cytoplasmic organelles, CBs lack any phospholipid membrane which would separate their...
and are referred to as Cajal body specific RNAs. This is in contrast to the majority of C/D box or H/ACA box snoRNAs which localise to the nucleolus. These Cajal body specific RNAs and are proposed to be involved in the modification of RNA polymerase II transcribed spliceosomal RNAs U1, U2, U4, U5 and U12. Not all snoRNAs that have been localised to Cajal bodies are composite C/D and H/ACA box snoRNAs.
Orphan snoRNAs
The targets for newly identified snoRNAs are predicted on the basis of sequence complementarity between putative target RNAs and the antisense elements or recognition loops in the snoRNA sequence. However, there are an increasing number of 'orphan' guides without any known RNA targets, which suggests that there might be more proteins or transcripts involved in rRNA than previously and/or that some snoRNAs have different functions not concerning rRNA. There is evidence that some of these orphan snoRNAs regulate alternatively spliced transcripts. For example, it appears that the C/D box snoRNA SNORD115Small nucleolar RNA SNORD115
SNORD115 is a non-coding RNA molecule known as a small nucleoloar RNA which usually functions in guiding the modification of other non-coding RNAs. This type of modifiying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis...
regulates the alternative splicing of the serotonin 2C receptor
5-HT2C receptor
The 5-HT2C receptor is a subtype of 5-HT receptor that binds the endogenous neurotransmitter serotonin . It is a G protein-coupled receptor that is coupled to Gq/G11 and mediates excitatory neurotransmission. HTR2C denotes the human gene encoding for the receptor, that in humans is located at the...
mRNA via a conserved region of complementarity.
Another C/D box snoRNA, SNORD116
Small nucleolar RNA SNORD116
SNORD116 is a non-coding RNA molecule which functions in the modification of other small nuclear RNAs . This type of modifiying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis...
, that resides in the same cluster as SNORD115 has been predicted to have 23 possible targets within protein coding genes using a bioinformatic approach. Of these a large fraction were found to be alternatively spliced, suggesting a role of SNORD116 in the regulation of alternative splicing.
Target modifications
The precise effect of the methylation and pseudouridylation modifications on the function of the mature RNAs is not yet known. The modifications do not appear to be essential but are known to subtly enhance the RNA folding and interaction with ribosomal proteins. In support of their importance, target site modifications are exclusively located within conserved and functionally important domains of the mature RNA and are commonly conserved amongst distant eukaryotes.- 2'-O-methylated ribose causes an increase in the 3'-endo conformation
- Pseudouridine (psi/Ψ) adds another option for H-bonding.
- Heavily methylated RNA is protected from hydrolysis. rRNA acts as a ribozyme by catalyzing its own hydrolysis and splicing.
Genomic organisation
The majority of vertebrate snoRNA genes are encoded in the introns of proteins involved in ribosome synthesis or translation, and are synthesized by RNA polymerase II, but snoRNAs can also be transcribed from their own promoters by RNA polymerase II or III.Imprinted loci
In the human genome there are at least two examples where C/D box snoRNAs are found in tandem repeats within imprinted loci. These two loci (14q32 on chromosome 14 and 15q11q13 on chromosome 15) have been extensively characterised and in both regions multiple snoRNAs have been found located within introns in clusters of closely related copies.In 15q11q13, five different snoRNAs have been identified (SNORD64
Small Nucleolar RNA SNORD64
SNORD64 is a non-coding RNA molecule which functions in the biogenesis of other small nuclear RNAs . This type of modifiying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis...
, SNORD107, SNORD108, SNORD109 (two copies), SNORD116
Small nucleolar RNA SNORD116
SNORD116 is a non-coding RNA molecule which functions in the modification of other small nuclear RNAs . This type of modifiying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis...
(29 copies) and SNORD115
Small nucleolar RNA SNORD115
SNORD115 is a non-coding RNA molecule known as a small nucleoloar RNA which usually functions in guiding the modification of other non-coding RNAs. This type of modifiying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis...
(48 copies). Loss of the 29 copies of SNORD116 (HBII-85) from this region has been identified as a cause of Prader-Willi syndrome
Prader-Willi syndrome
Prader–Willi syndrome is a rare genetic disorder in which seven genes on chromosome 15 are deleted or unexpressed on the paternal chromosome...
whereas gain of additional copies of SNORD115 has been linked to autism
Autism
Autism is a disorder of neural development characterized by impaired social interaction and communication, and by restricted and repetitive behavior. These signs all begin before a child is three years old. Autism affects information processing in the brain by altering how nerve cells and their...
.
Region 14q32 contains repeats of two snoRNAs SNORD113
Small nucleolar RNA SNORD113
Small nucleolar RNA SNORD113 is a small nucleolar RNA molecule which is located in the imprinted human 14q32 locus and may play a role in the evolution and/or mechanism of the epigenetic imprinting process....
(9 copies) and SNORD114 (31 copies) within the introns of a tissue-specific ncRNA transcript (MEG8
MEG8 (gene)
In molecular biology, Maternally expressed 8 , also known as MEG8 or Rian , is a long non-coding RNA. It is an imprinted gene, which is maternally expressed. It is expressed in the nucleus and is preferentially expressed in skeletal muscle....
). The 14q32 domain has been shown to share common genomic features with the imprinted 15q11-q13 loci and a possible role for tandem repeats of C/D box snoRNAs in the evolution or mechanism of imprinted loci has been suggested.
Other functions of snoRNA
snoRNAs can function as miRNAMirna
Mirna may refer to:geographical entities* Mirna , a river in Istria, Croatia* Mirna , a river in Slovenia, tributary of the river Sava* Mirna , a settlement in the municipality of Mirna in Southeastern Sloveniapeople...
s. It has been shown that human ACA45
Small Cajal body specific RNA 15
Small Cajal body specific RNA 15 is a small nucleolar RNA found in Cajal bodies and believed to be involved in the pseudouridylation of U1 spliceosomal RNA....
is a bona fide snoRNA that can be processed into a 21 nucleotides long mature miRNA by the RNAse III family endoribonuclease
RNase III
RNase III enzymes specifically bind to and cleave double-stranded RNA . There are three subdivisions, known as Class 1, 2, and 3.Prokaryotic ribonuclease III is an enzyme that digests double-stranded RNA. It is involved in the processing of ribosomal RNA precursors and of some mRNAs.* Class 1...
dicer
Dicer
Dicer is an endoribonuclease in the RNase III family that cleaves double-stranded RNA and pre-microRNA into short double-stranded RNA fragments called small interfering RNA about 20-25 nucleotides long, usually with a two-base overhang on the 3' end...
. This snoRNA product has previously been identified as mmu-miR-1839 and was shown to be processed independent of the other miRNA generating endoribonuclease
Endoribonuclease
A Endoribonuclease is a ribonuclease endonuclease. It cleaves either single-stranded or double-stranded RNA, depending on the enzyme. Example includes both single proteins like RNase III, RNase A, RNase T1, and RNase H, and also complexes of proteins like RNase P and the RNA-induced silencing...
drosha
Drosha
Drosha is a Class 2 RNase III enzyme responsible for initiating the processing of microRNA , or short RNA molecules naturally expressed by the cell that regulate a wide variety of other genes by interacting with the RNA-induced silencing complex to induce cleavage of complementary messenger RNA ...
. Bioinformatical
Bioinformatics
Bioinformatics is the application of computer science and information technology to the field of biology and medicine. Bioinformatics deals with algorithms, databases and information systems, web technologies, artificial intelligence and soft computing, information and computation theory, software...
analyses have revealed that putatively snoRNA-derived, miRNA-like fragments occur in different organisms.
Recently, it has been found that snoRNAs can have functions not related to rRNA. One such function is the regulation of alternative splicing
Alternative splicing
Alternative splicing is a process by which the exons of the RNA produced by transcription of a gene are reconnected in multiple ways during RNA splicing...
of the trans gene transcript, which is done by the snoRNA HBII-52
Small nucleolar RNA SNORD115
SNORD115 is a non-coding RNA molecule known as a small nucleoloar RNA which usually functions in guiding the modification of other non-coding RNAs. This type of modifiying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis...
, which is also known as SNORD115.
Recent discovery also show, the existence of snoRNA, microRNA, piRNA characteristics in a novel non-coding RNA: x-ncRNA and its biological implication in Homo sapiens.

