
Pseudoknot
Encyclopedia
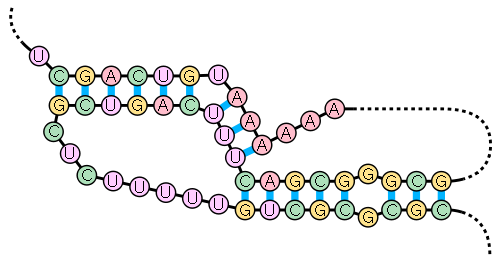
Nucleic acid secondary structure
The secondary structure of a nucleic acid molecule refers to the basepairing interactions within a single molecule or set of interacting molecules, and can be represented as a list of bases which are paired in a nucleic acid molecule....
containing at least two stem-loop
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions,...
structures in which half of one stem is intercalated between the two halves of another stem. The pseudoknot was first recognized in the turnip yellow mosaic virus in 1982. Pseudoknots fold into knot-shaped three-dimensional conformations but are not true topological knots
Knot (mathematics)
In mathematics, a knot is an embedding of a circle in 3-dimensional Euclidean space, R3, considered up to continuous deformations . A crucial difference between the standard mathematical and conventional notions of a knot is that mathematical knots are closed—there are no ends to tie or untie on a...
.
Prediction and identification
The structural configuration of pseudoknots does not lend itself well to bio-computational detection due to its context-sensitivity or “overlapping” nature. The base pairBase pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
ing in pseudoknots is not well nested; that is, base pairs occur that "overlap" one another in sequence position. This makes the presence of pseudoknots in RNA sequences more difficult to predict
Secondary structure prediction
Secondary structure prediction is a set of techniques in bioinformatics that aim to predict the secondary structures of proteins and nucleic acid sequences based only on knowledge of their primary structure...
by the standard method of dynamic programming
Dynamic programming
In mathematics and computer science, dynamic programming is a method for solving complex problems by breaking them down into simpler subproblems. It is applicable to problems exhibiting the properties of overlapping subproblems which are only slightly smaller and optimal substructure...
, which use a recursive scoring system to identify paired stems and consequently, most cannot detect non-nested base pairs. The newer method of stochastic context-free grammar
Stochastic context-free grammar
A stochastic context-free grammar is a context-free grammar in which each production is augmented with a probability...
s suffers from the same problem. Thus, popular secondary structure prediction methods like Mfold and Pfold will not predict pseudoknot structures present in a query sequence; they will only identify the more stable of the two pseudoknot stems.
It is possible to identify a limited class of pseudoknots using dynamic programming, but these methods are not exhaustive and scale worse as a function of sequence length than unpseudoknotted algorithms. The general problem of predicting lowest free energy structures with pseudoknots has been shown to be NP-complete
NP-complete
In computational complexity theory, the complexity class NP-complete is a class of decision problems. A decision problem L is NP-complete if it is in the set of NP problems so that any given solution to the decision problem can be verified in polynomial time, and also in the set of NP-hard...
.
Biological significance
Several important biological processes rely on RNA molecules that form pseudoknots. For example, the Telomerase RNA componentTelomerase RNA component
Telomerase RNA component, also known as TERC, is an RNA gene found in eukaryotes, that is a component of telomerase used to extend telomeres. Telomerase RNAs differ greatly in sequence and structure between vertebrates, ciliates and yeasts, but they share a 5' pseudoknot structure close to the...
contains a pseudoknot that is critical for activity.
Several viruses use a pseudoknot structure to form a tRNA-like motif to infiltrate the host cell. The pseudoknot region of RNase P
RNase P
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA...
is one of the most conserved elements in all of evolution.
RNA molecules with extensive tertiary structure often have substantial pseudoknots.

