Unfolded protein response
Encyclopedia
The unfolded protein response (UPR) is a cellular stress response related to the endoplasmic reticulum
. It is a stress response that has been found to be conserved between all mammal
ian species, as well as yeast
and worm organisms. This article focuses on the mammalian response.
The UPR is activated in response to an accumulation of unfolded or misfolded protein
s in the lumen
of the endoplasmic reticulum. In this scenario, the UPR has two primary aims: initially to restore normal function of the cell by halting protein translation
and activate the signaling pathways that lead to increasing the production of molecular chaperones involved in protein folding
. If these objectives are not achieved within a certain time lapse or the disruption is prolonged, the UPR aims to apoptosis
.
, RNA
, polypeptide) to the ER membrane. Once the sequence has “docked”, the protein continues translation, with the resultant strand being fed through the polypeptide translocator directly into the ER. Protein folding commences as soon as the polypeptide enters to the luminal environment, even as translation of the remaining polypeptide continues.
The ER is capable of recognising malfolding proteins without causing disruption to the functioning of the ER. The aforementioned sugar molecule remains the means by which the cell monitors protein folding, as the malfolding protein becomes characteristically devoid of glucose residues, targeting it for identification and re-glycosylation by the enzyme UGGT
(UGT-glucose:glycoprotein glucosyltransferase)1. If this fails to restore the normal folding process, exposed hydrophobic residues of the malfolded protein are bound by the protein glucose regulate protein 78 (Grp78), a heat shock protein 70kDa family member2 that prevents the protein from further transit and secretion3.
Where circumstances continue to cause a particular protein to malfold, the protein is recognised as posing a threat to the proper functioning of the ER, as they can aggregate to one another and accumulate. In such circumstances the protein is guided through endoplasmic reticulum-associated degradation (ERAD). The chaperone EDEM guides the retrotranslocation of the malfolded protein back into the cytosol in transient complexes with PDI and Grp784. Here it enters the ubiquitin-proteasome pathway, as it is tagged by multiple ubiquitin molecules, targeting it for degradation by cytosolic proteasomes.
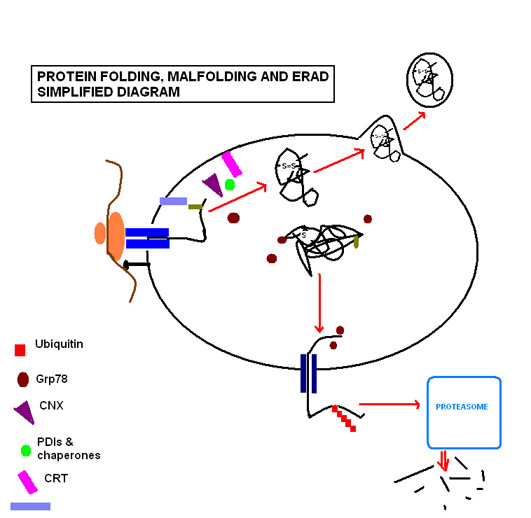 Successful protein folding requires a tightly controlled environment of substrates that include glucose to meet the metabolic energy requirements of the functioning molecular chaperones; calcium that is stored bound to resident molecular chaperones and; redox buffers that maintain the oxidising environment required for disulfide bond formation5.
Successful protein folding requires a tightly controlled environment of substrates that include glucose to meet the metabolic energy requirements of the functioning molecular chaperones; calcium that is stored bound to resident molecular chaperones and; redox buffers that maintain the oxidising environment required for disulfide bond formation5.
However where circumstances cause a more global disruption to protein folding that overwhelms the ER’s coping mechanisms, the UPR is activated.
Although this is traditionally the accepted model, doubts have been raised over its validity. It has been argued that the genetic and structural evidence supporting the model simply shows BiP dissociation to be merely correlated with Ire1 activation, rather than specifically causing it. An alternative model has been proposed, whereby unfolded proteins interact directly with the ER-lumenal domain of Ire1, causing oligomerization and transautophosphorylation.
Translation Attenuation and Cell Cycle Arrest by the PERK Receptor
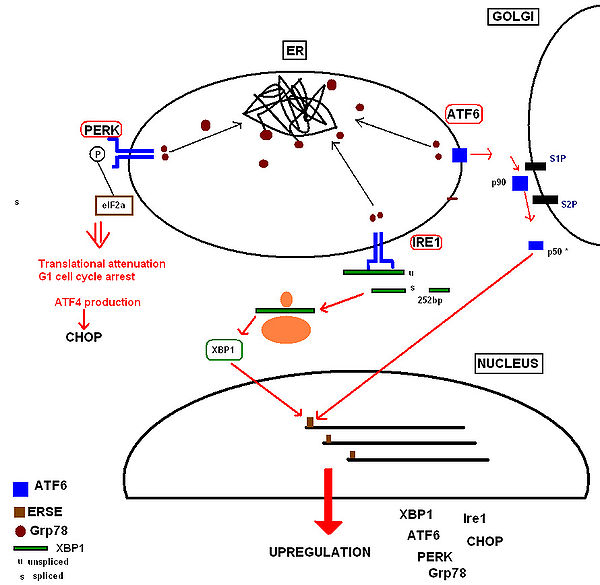
This occurs within minutes to hours of UPR activation to prevent further translational loading of the ER. PERK (protein kinase RNA-like endoplasmic reticulum kinase) activates itself by oligomerization and autophosphorylation of the free luminal domain. The activated cytosolic domain causes translational attenuation by directly phosphorylating the α subunit of the regulating initiator of the mRNA translation machinery, eIF26. This also produces translational attenuation of the protein machinery involved in running the cell cycle, producing cell cycle arrest in the G1 phase7.
 Increased Production of Proteins Involved in the Functions of the UPR
Increased Production of Proteins Involved in the Functions of the UPR
UPR activation also results in upregulation of proteins involved in chaperoning malfolding proteins, protein folding and ERAD, including further production of Grp78. Ultimately this increases the cell’s molecular mechanisms by which it can deal with the malfolded protein load. These receptor proteins have been identified as:
• Inositol-requiring kinase 18, whose free luminal domain activates itself by homodimerisation and transautophosphorylation9. The activated domain is able to activate the transcription factor XBP1 (X-box binding protein) mRNA (the mammalian equivalent of the yeast Hac1 mRNA) by cleavage and removal of a 252bp intron. The activated transcription factor upregulates UPR ‘stress genes’ by directly binding to stress element promoters in the nucleus10.
• ATF6 (activating transcription factor 6) is a basic leucine zipper transcription factor11.Upon Grp78 dissociation the entire 90kDa protein translocates to the Golgi, where it is cleaved by proteases to form an active 50kDa transcription factor12 that translocates to the nucleus. It binds to stress element promoters upstream of genes that are upregulated in the UPR 13.
The aim of these responses is to remove the accumulated protein load whilst preventing any further addition to the stress, so that normal function of the ER can be restored as soon as possible.
By binding with the protein TRAF2, Ire1 activates a JNK signaling pathway14, at which point human procaspase 4 is believed to cause apoptosis by activating downstream caspases.
Although PERK is recognised to produce a translational block, certain genes can bypass this block. An important example is that the proapoptotic protein CHOP (CCAAT/-enhancer-binding protein homologous protein),is upregulated downstream of the bZIP transcription factor ATF4 (activating transcription factor 4) and uniquely responsive to ER stress15. CHOP causes downregulation of the anti-apoptotic mitochondrial protein Bcl-216, favouring a pro-apoptotic drive at the mitochondria by proteins that cause mitochondrial damage, cytochrome c release and caspase 3 activation.
. Others are:
Endoplasmic reticulum
The endoplasmic reticulum is an organelle of cells in eukaryotic organisms that forms an interconnected network of tubules, vesicles, and cisternae...
. It is a stress response that has been found to be conserved between all mammal
Mammal
Mammals are members of a class of air-breathing vertebrate animals characterised by the possession of endothermy, hair, three middle ear bones, and mammary glands functional in mothers with young...
ian species, as well as yeast
Yeast
Yeasts are eukaryotic micro-organisms classified in the kingdom Fungi, with 1,500 species currently described estimated to be only 1% of all fungal species. Most reproduce asexually by mitosis, and many do so by an asymmetric division process called budding...
and worm organisms. This article focuses on the mammalian response.
The UPR is activated in response to an accumulation of unfolded or misfolded protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s in the lumen
Lumen (anatomy)
A lumen in biology is the inside space of a tubular structure, such as an artery or intestine...
of the endoplasmic reticulum. In this scenario, the UPR has two primary aims: initially to restore normal function of the cell by halting protein translation
Translation (genetics)
In molecular biology and genetics, translation is the third stage of protein biosynthesis . In translation, messenger RNA produced by transcription is decoded by the ribosome to produce a specific amino acid chain, or polypeptide, that will later fold into an active protein...
and activate the signaling pathways that lead to increasing the production of molecular chaperones involved in protein folding
Protein folding
Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil....
. If these objectives are not achieved within a certain time lapse or the disruption is prolonged, the UPR aims to apoptosis
Apoptosis
Apoptosis is the process of programmed cell death that may occur in multicellular organisms. Biochemical events lead to characteristic cell changes and death. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, and chromosomal DNA fragmentation...
.
Protein synthesis
The term protein folding incorporates all the processes involved in the production of a protein after the nascent polypeptides have become synthesized by the ribosomes. The proteins destined to be secreted or sorted to other cell organelles carry an N-terminal signal sequence that will interact with a signal recognition particle (SRP). The SRP will lead the whole complex (RibosomeRibosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
, RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
, polypeptide) to the ER membrane. Once the sequence has “docked”, the protein continues translation, with the resultant strand being fed through the polypeptide translocator directly into the ER. Protein folding commences as soon as the polypeptide enters to the luminal environment, even as translation of the remaining polypeptide continues.
Protein folding & Quality control
Protein folding steps involve a range of enzymes and molecular chaperones to coordinate and regulate reactions, in addition to a range of substrates required in order for the reactions to take place. The most important of these to note are N-linked glycosylation and disulfide bond formation. N-linked glycosylation occurs as soon as the protein sequence passes into the ER through the translocon, where it is glycosylated with a sugar molecule that forms the key ligand for the lectin molecules calreticulin (CRT) and calnexin (CNX)1. Favoured by the highly oxidising environment of the ER, Protein disulfide isomerases facilitate formation of disulfide bonds, which confer structural stability to the protein in order for it to withstand adverse conditions such as extremes of pH and degradative enzymes.The ER is capable of recognising malfolding proteins without causing disruption to the functioning of the ER. The aforementioned sugar molecule remains the means by which the cell monitors protein folding, as the malfolding protein becomes characteristically devoid of glucose residues, targeting it for identification and re-glycosylation by the enzyme UGGT
UGGT
UGGT, or -glucose:glycoprotein glucosyltransferase is a soluble enzyme resident in the lumen of the endoplasmic reticulum . The main function of UGGT is to recognize misfolded glycoproteins and transfer a glucose monomer to the terminal mannose on the glycoprotein...
(UGT-glucose:glycoprotein glucosyltransferase)1. If this fails to restore the normal folding process, exposed hydrophobic residues of the malfolded protein are bound by the protein glucose regulate protein 78 (Grp78), a heat shock protein 70kDa family member2 that prevents the protein from further transit and secretion3.
Where circumstances continue to cause a particular protein to malfold, the protein is recognised as posing a threat to the proper functioning of the ER, as they can aggregate to one another and accumulate. In such circumstances the protein is guided through endoplasmic reticulum-associated degradation (ERAD). The chaperone EDEM guides the retrotranslocation of the malfolded protein back into the cytosol in transient complexes with PDI and Grp784. Here it enters the ubiquitin-proteasome pathway, as it is tagged by multiple ubiquitin molecules, targeting it for degradation by cytosolic proteasomes.

However where circumstances cause a more global disruption to protein folding that overwhelms the ER’s coping mechanisms, the UPR is activated.
Initiation of the UPR
The molecular chaperone BiP/Grp78 has a range of functions within the ER. It maintains specific transmembrane receptor proteins involved in initiating of the downstream signalling of the UPR in an inactive state by binding to their luminal domains. An overwhelming load of misfolded proteins requires more of the available BiP/Grp78 to bind to the exposed hydrophobic regions of these proteins, and consequently BiP/Grp78 dissociates from these receptors sites to meet this requirement. Dissociation from the intracellular receptor domains allows them to become active.Although this is traditionally the accepted model, doubts have been raised over its validity. It has been argued that the genetic and structural evidence supporting the model simply shows BiP dissociation to be merely correlated with Ire1 activation, rather than specifically causing it. An alternative model has been proposed, whereby unfolded proteins interact directly with the ER-lumenal domain of Ire1, causing oligomerization and transautophosphorylation.
Functions of the UPR
The initial phases of UPR activation have two key roles:Translation Attenuation and Cell Cycle Arrest by the PERK Receptor
This occurs within minutes to hours of UPR activation to prevent further translational loading of the ER. PERK (protein kinase RNA-like endoplasmic reticulum kinase) activates itself by oligomerization and autophosphorylation of the free luminal domain. The activated cytosolic domain causes translational attenuation by directly phosphorylating the α subunit of the regulating initiator of the mRNA translation machinery, eIF26. This also produces translational attenuation of the protein machinery involved in running the cell cycle, producing cell cycle arrest in the G1 phase7.

UPR activation also results in upregulation of proteins involved in chaperoning malfolding proteins, protein folding and ERAD, including further production of Grp78. Ultimately this increases the cell’s molecular mechanisms by which it can deal with the malfolded protein load. These receptor proteins have been identified as:
• Inositol-requiring kinase 18, whose free luminal domain activates itself by homodimerisation and transautophosphorylation9. The activated domain is able to activate the transcription factor XBP1 (X-box binding protein) mRNA (the mammalian equivalent of the yeast Hac1 mRNA) by cleavage and removal of a 252bp intron. The activated transcription factor upregulates UPR ‘stress genes’ by directly binding to stress element promoters in the nucleus10.
• ATF6 (activating transcription factor 6) is a basic leucine zipper transcription factor11.Upon Grp78 dissociation the entire 90kDa protein translocates to the Golgi, where it is cleaved by proteases to form an active 50kDa transcription factor12 that translocates to the nucleus. It binds to stress element promoters upstream of genes that are upregulated in the UPR 13.
The aim of these responses is to remove the accumulated protein load whilst preventing any further addition to the stress, so that normal function of the ER can be restored as soon as possible.
Initiating Apoptosis
In conditions of prolonged stress, the goal of the UPR changes from being one that promotes cellular survival to one that commits the cell to a pathway of apoptosis. Proteins downstream of all 3 UPR receptor pathways have been identified as having pro-apoptotic roles. However, the point at which the ‘apoptotic switch’ is activated has not yet been determined, but it is a logical consideration that this should be beyond a certain time period in which resolution of the stress has not been achieved. The 2 principal UPR receptors involved are Ire1 and PERK.By binding with the protein TRAF2, Ire1 activates a JNK signaling pathway14, at which point human procaspase 4 is believed to cause apoptosis by activating downstream caspases.
Although PERK is recognised to produce a translational block, certain genes can bypass this block. An important example is that the proapoptotic protein CHOP (CCAAT/-enhancer-binding protein homologous protein),is upregulated downstream of the bZIP transcription factor ATF4 (activating transcription factor 4) and uniquely responsive to ER stress15. CHOP causes downregulation of the anti-apoptotic mitochondrial protein Bcl-216, favouring a pro-apoptotic drive at the mitochondria by proteins that cause mitochondrial damage, cytochrome c release and caspase 3 activation.
Chemical inducers of the UPR
UPR inducers most notably include tunicamycinTunicamycin
Tunicamycin is mixture of homologous nucleoside antibiotics that inhibits the UDP-HexNAc: polyprenol-P HexNAc-1-P family of enzymes. In eukaryotes, this includes the enzyme GlcNAc phosphotransferase , which catalyzes the transfer of N-actelyglucosamine-1-phosphate from UDP-N-acetylglucosamine to...
. Others are:
- thapsigarginThapsigarginThapsigargin is non-competitive inhibitor of a class of enzymes known by the acronym SERCA, which stands for sarco / endoplasmic reticulum Ca2+ ATPase. Structurally, thapsigargin is classified as a sesquiterpene lactone, and is extracted from a plant, Thapsia garganica. It is a tumor promoter in...
Leads to ER Ca+2 depletion due to inhibition of the Sarco/Endoplasmic Reticulum Ca2+-ATPase (SERCA). - A23187A23187A23187 is a mobile ion-carrier that forms stable complexes with divalent cations . A23187 is also known as Calcimycin, Calcium Ionophore, Antibiotic A23187 and Calcium Ionophore A23187...
- 2-deoxyglucose
- dithiothreitolDithiothreitolDithiothreitol is the common name for a small-molecule redox reagent known as Cleland's reagent. DTT's formula is C4H10O2S2 and the molecular structure of its reduced form is shown at the right; its oxidized form is a disulfide-bonded 6-membered ring . Its name derives from the four-carbon...
Reduce the disulfide bridges of proteins. The denatured proteins accumulated inside the ER.
Further reading
- Blond-Elguindi, S., Cwiria, SE., Dower, WJ., Lipshutz, RJ., Sprang, SR., Sambrook, JF., Gething, MH (1993) Cell 75: 717-728
- Brewer, J., Diehl, J. (2000) Proc Natl Acad USA 97 (23): 12625-30
- Chen, X., Shen, J., Prywes, R. (2002) J Biol Chem 277 (15): 13045-53
- Cox, JS., Shamu, CE., Walter, P. (1993) Cell 73 (6): 1197-1206
- Hammond, C., Braakman, I., Helenius, A. 1994 PNAS 91: 913-917
- Harding, H. P., Zhang, Y., Ron, D. (1999) Nature 397 271-4
- Lee, A-H., Iwakoshi, N., Anderson, K., Glimcher, L. (2003) Proc Natl Acad Sci USA 100 (17) 9946-51
- Lee, AS (1987) Trends Biochem Sci 12 20-23
- Machamer, CE., Doms, RW., Bole, DG,. Helenius, A., Rose, JK. (1990) J Biol Chem 265 (12) 6879-6883
- McCullough, K., Martindale, J., Klotz, L., Aw, T., Holbrook, N (2001) Mol Cell Biol 21: 1249-1259
- Molinari, M., Galli, C., Piccaluga, V., Pieren, M., Paganetti, P. (2002) J Cell Biol 158 (2) 247-257
- Mori, K., Ogawa, O., Kawahara, T., Yanagi, H., Yura, T. (2000) Proc Natl Acad Sci USA 97 4660-4665
- Urano, F., Wang, X., Bertolotti, A., Zhang, Y., Chung, P., Harding, H., Ron, D (2000) Science 287 (5453) 664-666
- Wang, X-Z., Lawson, B., Brewer, J. W., Zinszner, H., Sanjay, A., Mi, L., Boorstein, R., Kreibich, G., Hendershot, L., Ron., D. (1996) Mol Cell Biol 16 (8) 4273-80
- Welihinda, A. A., Kaufman, R. J. (1996) J Biol Chem 271 (30) 18181-7
- Yoshida, H., Haze, K., Yanagi, H., Yura, T., Mori, K. (1998) J Biol Chem 273 (50): 33741-9

