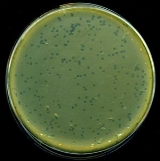
Lambda phage
Encyclopedia
Lambda
Lambda is the 11th letter of the Greek alphabet. In the system of Greek numerals lambda has a value of 30. Lambda is related to the Phoenician letter Lamed . Letters in other alphabets that stemmed from lambda include the Roman L and the Cyrillic letter El...
phage, coliphage λ) is a temperate
Temperate (virology)
In virology, temperate refers to the ability of some bacteriophages to display a lysogenic life cycle. Many temperate phages can integrate their genomes into their host bacterium's chromosome, together becoming a lysogen as the phage genome becomes a prophage...
bacteriophage
Bacteriophage
A bacteriophage is any one of a number of viruses that infect bacteria. They do this by injecting genetic material, which they carry enclosed in an outer protein capsid...
that infects Escherichia coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
.
Lambda phage is a virus
Virus
A virus is a small infectious agent that can replicate only inside the living cells of organisms. Viruses infect all types of organisms, from animals and plants to bacteria and archaea...
particle consisting of a head, containing double-stranded linear DNA as its genetic material, and a tail that can have tail fibers. The phage particle recognizes and binds to its host, E. coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
, causing DNA in the head of the phage to be ejected through the tail into the cytoplasm of the bacterial cell. Usually, a "lytic cycle
Lytic cycle
The lytic cycle is one of the two cycles of viral reproduction, the other being the lysogenic cycle. The lytic cycle is typically considered the main method of viral replication, since it results in the destruction of the infected cell...
" ensues, where the lambda DNA is replicated many times and the genes for head, tail and lysis proteins are expressed. This leads to assembly of multiple new phage particles within the cell and subsequent cell lysis
Lysis
Lysis refers to the breaking down of a cell, often by viral, enzymic, or osmotic mechanisms that compromise its integrity. A fluid containing the contents of lysed cells is called a "lysate"....
, releasing the cell contents, including virions that have been assembled, into the environment. However, under certain conditions the phage DNA may integrate itself into the host cell chromosome in the lysogenic pathway. In this state, the λ DNA is called a prophage
Prophage
A prophage is a phage genome inserted and integrated into the circular bacterial DNA chromosome. A prophage, also known as a temperate phage, is any virus in the lysogenic cycle; it is integrated into the host chromosome or exists as an extrachromosomal plasmid. Technically, a virus may be called...
and stays resident within the host's genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
without apparent harm to the host. The host can be termed a lysogen
Lysogen
A lysogen or lysogenic phage is a phage that can exist as a prophage within its host organism. A prophage is the phage DNA in its dormant state, and is either integrated into the host bacteria's chromosome or more rarely exists as a stable plasmid within the host cell...
when a prophage is present. The prophage is duplicated with every subsequent cell division of the host. The phage genes expressed in this dormant state code for protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s that repress expression of other phage genes (such as the structural and lysis genes) in order to prevent entry into the lytic cycle. These repressive proteins are broken down when the host cell is under stress, resulting in the expression of the repressed phage genes. Stress can be from starvation
Starvation
Starvation is a severe deficiency in caloric energy, nutrient and vitamin intake. It is the most extreme form of malnutrition. In humans, prolonged starvation can cause permanent organ damage and eventually, death...
, poisons (like antibiotics), or other factors that can damage or destroy the host. In response to stress, the activated prophage is excised from the DNA of the host cell by one of the newly expressed gene products and enters its lytic pathway.
Lambda phage was discovered by Esther Lederberg
Esther Lederberg
Esther Miriam Zimmer Lederberg was an American microbiologist and immunologist and pioneer of bacterial genetics...
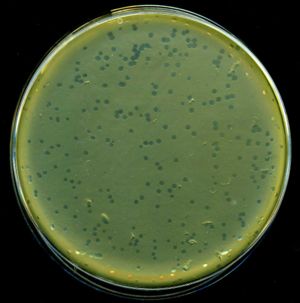
in 1950 when it was noticed that a colony produced by replica plating
Replica plating
In molecular biology and microbiology, replica plating is a technique in which one or more secondary Petri plates containing different solid selective growth media are inoculated with the same colonies of microorganisms from a primary plate , reproducing the original spatial...
was "nibbled and plaqued
Viral plaque
A viral plaque is a visible structure formed within a cell culture, such as bacterial cultures within some nutrient medium . The bacteriophage viruses replicate and spread, thus generating regions of cell destructions known as plaques....
". Lambda phage has been used heavily as a model organism
Model organism
A model organism is a non-human species that is extensively studied to understand particular biological phenomena, with the expectation that discoveries made in the organism model will provide insight into the workings of other organisms. Model organisms are in vivo models and are widely used to...
, and has been a rich source for useful tools in microbial genetics
Microbial genetics
Microbial genetics is a subject area within microbiology and genetic engineering. It studies the genetics of very small organisms. This involves the study of the genotype of microbial species and also the expression system in the form of phenotypes.It also involves the study of genetic processes...
, and later in molecular genetics
Molecular genetics
Molecular genetics is the field of biology and genetics that studies the structure and function of genes at a molecular level. The field studies how the genes are transferred from generation to generation. Molecular genetics employs the methods of genetics and molecular biology...
. Uses include its application as a vector for the cloning of recombinant DNA
Recombinant DNA
Recombinant DNA molecules are DNA sequences that result from the use of laboratory methods to bring together genetic material from multiple sources, creating sequences that would not otherwise be found in biological organisms...
; the use of its site-specific recombinase (int) for the shuffling of cloned DNAs by the gateway method
Gateway Technology
The Gateway cloning System, invented and commercialized by Invitrogen since the late 1990s, is a molecular biology method that enables researchers to efficiently transfer DNA-fragments between plasmids using a proprietary set of recombination sequences, the "Gateway att" sites, and two proprietary...
; and the application of its Red operon
Operon
In genetics, an operon is a functioning unit of genomic DNA containing a cluster of genes under the control of a single regulatory signal or promoter. The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo trans-splicing to create...
, including the proteins Red alpha (also called 'exo'), beta and gamma in the DNA engineering method called recombineering
Recombineering
-Definition of Recombineering:Recombineering is a genetic and molecular biology technique based on homologous recombination systems, as opposed to the older/more common method of using restriction enzymes and ligases to cut and glue DNA sequences. It has been developed in E...
.
Anatomy
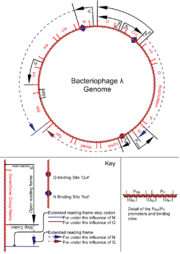
The virus particle consists of a head and a tail that can have tail fibers. The head contains 48,490 base pairs of double-stranded, linear DNA, with 12-base single-stranded segments at both 5' ends. These two single-stranded segments are the "sticky ends" of what is called the cos site. The cos site circularizes the DNA in the host cytoplasm. In its circular form, the phage genome therefore is 48,502 base pairs in length. The prophage exists as a linear section of DNA inserted into the host chromosome.Lifecycle
Infection
- Bacteriophage Lambda binds to the target E. coli cell, the J protein in the tail tip interacting with the lamB gene product of E. coli, a porinPorin (protein)Porins are beta barrel proteins that cross a cellular membrane and act as a pore through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules...
molecule which is part of the maltoseMaltoseMaltose , or malt sugar, is a disaccharide formed from two units of glucose joined with an αbond, formed from a condensation reaction. The isomer "isomaltose" has two glucose molecules linked through an α bond. Maltose is the second member of an important biochemical series of glucose chains....
operon. - The linear phage genome is injected past the cell outer membrane.
- The DNA passes through a separate sugar transport protein (ptsG) in the inner membrane, and immediately circularises using the cos sites, 12-base G-C rich cohesive "sticky ends". The single-stranded nicks are ligated by host DNA ligaseDNA ligaseIn molecular biology, DNA ligase is a specific type of enzyme, a ligase, that repairs single-stranded discontinuities in double stranded DNA molecules, in simple words strands that have double-strand break . Purified DNA ligase is used in gene cloning to join DNA molecules together...
. - Host DNA gyraseDNA gyraseDNA gyrase, often referred to simply as gyrase, is an enzyme that relieves strain while double-stranded DNA is being unwound by helicase. This causes negative supercoiling of the DNA...
puts negative supercoils in the circular chromosome, causing A-T rich regions to unwind and drive transcription. - Transcription starts from the constitutive PL, PR and PR' promoters producing the 'immediate early' transcripts. Initially these express the N and cro genes, producing N, Cro and a short inactive protein.
- Cro binds to OR3 preventing access to the PRM promoter preventing expression of the cI gene. N binds to the two Nut (N utilisation) sites, one in the N gene in the PL reading frame, and one in the cro gene in the PR reading frame.
- The N protein is an antiterminator, and functions to extend the reading frames to which it is bound. When RNA polymeraseRNA polymeraseRNA polymerase is an enzyme that produces RNA. In cells, RNAP is needed for constructing RNA chains from DNA genes as templates, a process called transcription. RNA polymerase enzymes are essential to life and are found in all organisms and many viruses...
transcribes these regions, it recruits the N and forms a complex with several host Nus proteins. This complex skips through most termination sequences. The extended transcripts (the 'late early' transcripts) include the N and cro genes along with cII and cIII genes, and xis, int, O, P and Q genes discussed later. - The cIII protein acts to protect the cII protein from proteolysis by FtsH (a membrane-bound essential E. coli protease) by acting as a competitive inhibitor. This inhibition can induce a bacteriostatic state, which favours lysogeny. cIII also directly stabilises the cII protein. On initial infection, the stability of cII determines the lifestyle of the phage; stable cII will lead to the lysogenic pathway, whereas if cII is degraded the phage will go into the lytic pathway. Low temperature, starvation of the cells and high multiplicity of infection (MOI) are known to favor lysogeny (see later discussion).
N antitermination
This occurs without the N protein interacting with the DNA; the protein instead binds to the freshly transcribed mRNA. Nut sites contain 3 conserved "boxes," of which only BoxB is essential.- The boxB RNA sequences are located close to the 5' end of the pL and pR transcripts. When transcribed, each sequence forms a hairpin loop structure that the N protein can bind to.
- N protein binds to boxB in each transcript, and contacts the transcribing RNA polymerase via RNA looping. The N-RNAP complex is stabilized by subsequent binding of several host Nus (N utilisation substance) proteins (which include transcription termination/antitermination factors and, bizarrely, a ribosome subunit).
- The entire complex (including the bound Nut site on the mRNA) continues transcription, and can skip through termination sequences.
Lytic life cycle
This is the lifecycle that the phage follows following most infections, where the cII protein does not reach a high enough concentration due to degradation, so does not activate its promoters.- The 'late early' transcripts continue being written, including xis, int, Q and genes for replication of the lambda genome (OP). Cro dominates the repressor site (see "Repressor" section), repressing synthesis from the PRM promoter.
- The O and P proteins initiate replication of the phage chromosome (see "Lytic Replication").
- Q, another antiterminator, binds to Qut sites.
- Transcription from the PR' promoter can now extend to produce mRNA for the lysis and the head and tail proteins.
- Structural proteins and phage genomes self assemble into new phage particles.
- Products of the lysis genes S,R, Rz and Rz1 cause cell lysis. S is a holin, a small membrane protein that, at a time determined by the sequence of the protein, suddenly makes holes in the membrane. R is an endolysinEndolysinEndolysin is a generic term describing an enzyme that degrades the bacterial peptidoglycan cell wall, resulting in lysis of the bacterial cell....
, an enzyme that escapes through the S holes and cleaves the cell wall. Rz and Rz1 are membrane proteins that form a complex that somehow destroys the outer membrane, after the endolysin has degraded the cell wall. For wild-type lambda, lysis occurs at about 50 minutes after the start of infection and releases around 100 virions.
Rightward transcription
Rightward transcription expresses the O, P and Q genes. O and P are responsible for initiating replication, and Q is another antiterminator which allows the expression of head, tail and lysis genes from PR’.Lytic replication
- For the first few replication cycles, the lambda genome undergoes θ replicationTheta structureA Theta structure is an intermediate structure formed during the replication of a circular DNA molecule , two replication forks can proceed independently around the DNA ring and when viewed from above it resembles the Greek letter "theta"...
(circle-to-circle). - This is initiated at the ori site located in the O gene. O protein binds the ori site, and P protein binds the DnaB subunit of the host replication machinery as well as binding O. This effectively commandeers the host DNA polymerase.
- Soon, the phage switches to a rolling-circle type of replication similar to that used by phage M13. The DNA is nicked and the 3’ end serves as a primer. Notably, this doesn’t release single copies of the phage genome, but rather one long molecule with many copies of the genome: a concatemer.
- These concatemers are cleaved at their cos sites as they are packaged. Packaging cannot occur from circular phage DNA, only from concatomeric DNA.
Q antitermination
Q is similar to N in its effect: Q binds to RNA polymerase in Qut sites and the resulting complex can ignore terminators, however the mechanism is very different; the Q protein first associates with a DNA sequence rather than an mRNA sequence.
- The Qut site is very close to the PR’ promoter, close enough that the σ factor has not been released from the RNA polymerase holoenzyme. Part of the Qut site resembles the -10 Pribnow boxPribnow boxThe Pribnow box is the sequence TATAAT of six nucleotides that is an essential part of a promoter site on DNA for transcription to occur in bacteria...
, causing the holoenzyme to pause. - Q protein then binds and displaces part of the σ factor and transcription re-initiates.
- The head and tail genes are transcribed and the corresponding proteins self-assemble.
Leftward transcription
Leftward transcription expresses the gam, red, xis and int genes. Gam and red proteins are involved in recombination. Gam is also important in that it inhibits the host RecBCD nuclease from degrading the 3’ ends in rolling circle replication. Int and xis are integration and excision proteins which are vital to lysogeny.xis and int regulation of insertion and excision
- xis and int are found on the same piece of mRNA, so approximately equal concentrations of xis and int proteins are produced. This results (initially) in the excision of any inserted genomes from the host genome.
- The mRNA from the PL promoter forms a stable secondary structure with a bobby pin loop in the sib section of the mRNA. This targets the 3' (sib) end of the mRNA for RNAaseIII degradation, which results in a lower effective concentration of int mRNA than xis mRNA (as the int cistron is nearer to the sib sequence than the xis cistron is to the sib sequence), so a higher concentrations of xis than int is observed.
- Higher concentrations of xis than int result in no insertion or excision of phage genomes, the evolutionarily favoured action - leaving any pre-insterted phage genomes inserted (so reducing competition) and preventing the insertion of the phage genome into the genome of a doomed host.
Lysogenic (or lysenogenic) life cycle
This is the lifecycle that the phage follows after a small number of infections in specific conditions, where the cII protein reaches a high enough concentration due to stabilisation and lack of degradation, and so activates its promoters.- The 'late early' transcripts continue being written, including xis, int, Q and genes for replication of the lambda genome.
- The stabilized cII acts to promote transcription from the PRE, PI and Pantiq promoters.
- The Pantiq promoter produces antisense mRNA to the Q gene message of the PR promoter transcript, thereby switching off Q production. The PRE promoter produces antisense mRNA to the cro section of the PR promoter transcript, turning down cro production, and has a transcript of the cI gene. This is expressed, turning on cI repressor production. The PI promoter expresses the int gene, resulting in high concentrations of int protein. This int protein integrates the phage DNA into the host chromosome (see "Prophage Integration").
- No Q results in no extension of the PR' promoter's reading frame, so no lytic or structural proteins are made. Elevated levels of int (much higher than that of xis) result in the insertion of the lambda genome into the hosts genome (see diagram). Production of cI leads to the binding of cI to the OR1 and OR2 sites in the PR promoter, turning off cro and other early gene expression. cI also binds to the PL promoter, turning off transcription there too.
- Lack of cro leaves the OR3 site unbound, so transcription from the PRM promoter may occur, maintaining levels of cI.
- Lack of transcription from the PL and PR promoters leads to no further production of cII and cIII.
- As cII and cIII concentrations decrease, transcription from the Pantiq, PRE and PI stop being promoted since they are no longer needed.
- Only the PRM and PR' promoters are left active, the former producing cI protein and the latter a short inactive transcript. The genome remains inserted into the host genome in a dormant state.
Prophage integration
The integration of phage λ takes place at a special attachment site in the bacterial and phage genomes, called attλ. The sequence of the bacterial att site is called attB, between the gal and bio operons, and consists of the parts B-O-B', whereas the complementary sequence in the circular phage genome is called attP and consists of the parts P-O-P'. The integration itself is a sequential exchange (see genetic recombinationGenetic recombination
Genetic recombination is a process by which a molecule of nucleic acid is broken and then joined to a different one. Recombination can occur between similar molecules of DNA, as in homologous recombination, or dissimilar molecules, as in non-homologous end joining. Recombination is a common method...
) via a Holliday junction
Holliday junction
A Holliday junction is a mobile junction between four strands of DNA. The structure is named after Robin Holliday, who proposed it in 1964 to account for a particular type of exchange of genetic information he observed in yeast known as homologous recombination...
and requires both the phage protein Int and the bacterial protein IHF (integration host factor). Both Int and IHF bind to attP and form an intasome, a DNA-protein-complex designed for site-specific recombination
Site-specific recombination
Site-specific recombination, also known as conservative site-specific recombination, is a type of genetic recombination in which DNA strand exchange takes place between segments possessing only a limited degree of sequence homology...
of the phage and host DNA. The original B-O-B' sequence is changed by the integration to B-O-P'-phage DNA-P-O-B'. The phage DNA is now part of the host's genome.
Maintenance of lysogeny
- Lysogeny is maintained solely by cI. cI represses transcription from PL and PR while upregulating and controlling its own expression from PRM. It is therefore the only protein expressed by lysogenic phage.
- This is coordinated by the PL and PR operators. Both operators have three binding sites for cI: OL1, OL2, and OL3 for PL, and OR1, OR2 and OR3 for PR.
- cI binds most favorably to OR1; binding here inhibits transcription from PR. As cI easily dimerises, the binding of cI to OR1 greatly increases the affinity of the binding of cI to OR2, and this happens almost immediately after OR1 binding. This activates transcription in the other direction from PRM, as the N terminal domain of cI on OR2 tightens the binding of RNA polymerase to PRM and hence cI stimulates its own transcription. When it is present at a much higher concentration, it also binds to OR3, inhibiting transcription from PRM, thus regulating its own levels in a negative feedback loop.
- cI binding to the PL operator is very similar, except that it has no direct effect on cI transcription. As an additional repression of its own expression, however, cI dimers bound to OR3 and OL3 bend the DNA between them to tetramerise.
- The presence of cI causes immunity to superinfection by other lambda phages, as it will inhibit their PL and PR promoters.
Induction
The classic induction of a lysogen involved irradiating the infected cells with UV light. Any situation where a lysogen undergoes DNA damage or the SOS response of the host is otherwise stimulated leads to induction.- The host cell, containing a dormant phage genome, experiences DNA damage due to a high stress environment, and starts to undergo the SOS responseSOS responseThe SOS response is a global response to DNA damage in which the cell cycle is arrested and DNA repair and mutagenesis are induced. The SOS uses the RecA protein . The RecA protein, stimulated by single-stranded DNA, is involved in the inactivation of the LexA repressor thereby inducing the response...
. - RecA (a cellular protein) detects DNA damage and becomes activated. It is now RecA*, a highly specific co-protease.
- Normally RecA* binds LexA (a transcriptionTranscription (genetics)Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
repressor), activating LexA auto-protease activity,which destroys LexA repressor allowing production of DNA repairDNA repairDNA repair refers to a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as UV light and radiation can cause DNA damage, resulting in as many as 1...
proteins. In lysogenic cells this response is hijacked, and RecA* stimulates cI autocleavage. This is because cI mimics the structure of LexA at the autocleavage site. - Cleaved cI can no longer dimerise, and loses its affinity for DNA binding.
- The PR and PL promoters are no longer repressed and switch on, and the cell returns to the lytic sequence of expression events (note that cII is not stable in cells undergoing the SOS response). There is however one notable difference.
Control of phage genome excision in induction
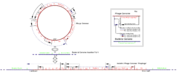
- The phage genome is still inserted in the host genome and needs excision for DNA replication to occur. The sib section beyond the normal PL promoter transcript is, however, no longer included in this reading frame (see diagram).
- No sib domain on the PL promoter mRNA results in no hairpin loop on the 3' end, and the transcript is no longer targeted for RNAaseIII degradation.
- The new intact transcript has one copy of both xis and int, so approximately equal concentrations of xis and int proteins are produced.
- Equal concentrations of xis and int result in the excision of the inserted genome from the host genome for replication and later phage production.
Protein function overview
cro; (Control of Repressor's Operator) Transcription inhibitor, binds OR3, OR2 and OR1 (affinity OR3 > OR2 = OR1, i.e. preferentially binds OR3). At low concentrations blocks the pRM promoter (preventing cI production). At high concentrations downregulates its own production through OR2 and OR1 binding. No cooperative binding (c.f. below for cI binding)cI; (Clear 1) Transcription inhibitor, binds OR1, OR2 and OR3 (affinity OR1 > OR2 = OR3, i.e. preferentially binds OR1). At low concentrations blocks the pR promoter (preventing cro production). At high concentrations downregulates its own production through OR3 binding. Binding of cI at OR1 stimulates an almost simultaneous cI binding to OR2 via cooperative binding (via cI C terminal domain interactions) N terminal domain of cI on OR2 tightens the binding of RNA polymerase holoenzyme complex to pRM and hence stimulate its own transcription. Repressor also inhibits transcription from the pL promoter. Susceptible to cleavage by RecA* in cells undergoing the SOS response.
cII; (Clear 2) Transcription activator. Activates transcription from the pAQ, pRE and pI promoters. Low stability due to susceptibility to cellular HflB (FtsH) proteases (especially in healthy cells and cells undergoing the SOS response).
cIII;(Clear 3) HflB (FtsH) binding protein, protects cII from degradation by proteases.
N; (aNtiterminator) RNA binding protein and RNA polymerase cofactor
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound that is bound to a protein and is required for the protein's biological activity. These proteins are commonly enzymes, and cofactors can be considered "helper molecules" that assist in biochemical transformations....
, binds RNA (at Nut sites) and transfers onto the nascent RNApol that just transcribed the nut site. This RNApol modification prevents its recognition of termination sites, so normal RNA polymerase termination signals are ignored and RNA synthesis continues into distal phage genes.
Q; DNA binding protein and RNApol cofactor, binds DNA (at Qut sites) and transfers onto the initiating RNApol. This RNApol modification alters its recognition of termination sequences, so normal ones are ignored; special Q termination sequences some 20,000 bp away are effective.
xis; (eXcISion) excisionase
Excisionase
In molecular biology, excisionase is a bacteriophage protein encoded by the Xis gene. It is involved in excisive recombination by regulating the assembly of the excisive intasome and by inhibiting viral integration. It adopts an unusual winged-helix structure in which two alpha helices are packed...
and Int protein regulator, manages excision and insertion of phage genome into the host's genome.
int; (INTegration) Int protein, manages insertion of phage genome into the host's genome. In Conditions of low int concentration there is no effect. If xis is low in concentration and int high then this leads to the insertion of the phage genome. If xis and int have high (and approximately equal) concentrations this leads to the excision of phage genomes from the host's genome.
A, B, C, D, E, F, Z, U, V, G, T, H, M, L, K, I, J [Shown on diagram as head and tail, A-F code for phage head genes, Z-J code for phage tail genes. The order shown here is as found on the genome, reading in a clockwise direction]; structural proteins, self assemble with the phage genome into daughter phage particles.
S, R [Shown on diagram as lysis. The order shown here is as found on the genome, reading in a clockwise direction]; cause the host cell to undergo lysis at high enough concentrations.
OP [Shown on diagram as O replication P]; DNA replication functions, promotes the specific replication of only the phage genome.
sib [not a protein, but a vital conserved DNA sequence]; Forms a stable hairpin loop structure in transcribed mRNA beyond int. Attracts degradation of mRNA by RNAaseIII.
attP [not a protein, but a conserved DNA sequence]; point of action of Int and Xis in integration and excision of the phage genome into the host's genome. Corresponding attB found in the host's genome at the point of insertion.
Repressor
The repressorRepressor
In molecular genetics, a repressor is a DNA-binding protein that regulates the expression of one or more genes by binding to the operator and blocking the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes. This blocking of expression is called...
found in the phage lambda is a notable example of the level of control possible over gene expression by a very simple system. It forms a 'binary switch' with two genes under mutually exclusive expression, as discovered by Barbara J. Meyer
Barbara J. Meyer
Barbara J. Meyer is a biologist, noted for her pioneering research on lambda phage, a virus that infects bacteria; discovery of the master control gene involved in sex determination; and studies of gene regulation, particularly dosage compensation.-Biography:...
.
The lambda repressor gene system consists of (from left to right on the chromosome):
- cI gene
- OR3
- OR2
- OR1
- cro gene
The lambda repressor is a dimer also known as the cI protein
CI protein
cI protein , also known as the lambda repressor, is the sole protein expressed in the lysogenic state of Lambda phage. cI turns off transcription at the phage's L and R promoters as well as any other invading lambda phages into the host cell. It is a transcription factor and contains the common...
. It binds DNA in the helix-turn-helix binding motif. It regulates the transcription of the cI protein and the Cro protein.
The life cycle of lambda phages is controlled by cI and Cro proteins. The lambda phage will remain in the lysogenic state if cI proteins predominate, but will be transformed into the lytic cycle if cro proteins predominate.
The cI dimer may bind to any of three operators, OR1, OR2, and OR3, in the order OR1 = OR2 > OR3.
Binding of a cI dimer to OR1 enhances binding of a second cI dimer to OR2, an effect called cooperativity
Cooperativity
Cooperativity is a phenomenon displayed by enzymes or receptors that have multiple binding sites where the affinity of the binding sites for a ligand is increased, positive cooperativity, or decreased, negative cooperativity, upon the binding of a ligand to a binding site...
. Thus, OR1 and OR2 are almost always simultaneously occupied by cI. However, this does not increase the affinity between cI and OR3, which will be occupied only when the cI concentration is high. This cooperative action is shown by the relative affinity of the repressor for the native sequences individually, which is OR1 > OR2 = OR3; different from the actual order of binding.
- In the absence of cI proteins, the cro gene may be transcribed.
- In the presence of cI proteins, only the cI gene may be transcribed.
- At high concentration of cI, transcriptions of both genes are repressed.
Lytic or lysogenic?
The geneGene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
regulatory circuitry of phage λ is among the best-understood circuits at the mechanistic level. This circuitry involves several interesting regulatory behaviors. An infected cell undergoes a decision between two alternative pathways, the lytic and lysogenic pathways. If the latter is followed, the lysogenic state is established and maintained. While this state is highly stable, it can switch to the lytic pathway in the process of prophage induction, which occurs when the host SOS response is triggered by DNA damage.
An important distinction here is that between the two decisions; lysogeny and lysis on infection, and continuing lysogeny or lysis from a prophage. The latter is determined solely by the activation of RecA in the SOS response of the cell, as detailed in the section on induction. The former will also be affected by this; a cell undergoing an SOS response will always by lysed, as no cI protein will be allowed to build up. However, the initial lytic/lysogenic decision on infection is also dependent on the cII and cIII proteins.
Simplistically, in cells with sufficient nutrients, protease activity is high, which breaks down cII. This leads to the lytic lifestyle. In cells with limited nutrients, protease activity is low, making cII stable. This leads to the lysogenic lifestyle. cIII appears to stabilize cII, both directly and by acting as a competitive inhibitor to the relevant proteases. This means that a cell "in trouble", i.e. lacking in nutrients and in a more dormant state, is more likely to lysogenise. This would be selected for because the phage can now lie dormant in the bacterium until it falls on better times, and so the phage can create more copies of itself with the additional resources available and with the more likely proximity of further infectable cells.
Realistically, a full biophysical model for lambda's lysis-lysogeny decision remains to be developed. Computer modeling and simulation suggest that random processes during infection drive the selection of lysis or lysogeny within individual cells. However, recent experiments suggest that physical differences among cells, that exist prior to infection, predetermine whether a cell will lyse or become a lysogen.
Other Related Areas of Research
Lambda phage has been of major importance in many other areas of research, such as specialized Transduction, GalactosemiaGalactosemia
Galactosemia is a rare genetic metabolic disorder that affects an individual's ability to metabolize the sugar galactose properly. Although the sugar lactose can metabolize to galactose, galactosemia is not related to and should not be confused with lactose intolerance...
, Maltophilia, and antibiotic drug resistance to name a few.
See also
- molecular weight size markerMolecular weight size markerA molecular weight size marker is used to identify the approximate size of a molecule run on a gel, using the principle that molecular weight is inversely proportional to migration rate through a gel matrix...
- Time-lapse microscopy video from MIT showing both lysis and lysogeny by phage lambda.
Further reading
- James Watson, Tania Baker, Stephen Bell, Alexander Gann, Michael Levine, Richard Losick " Molecular Biology of the Gene (International Edition)" - 6th Edition
- Mark PtashneMark PtashneMark Ptashne is a molecular biologist and violinist. He currently holds the Ludwig Chair of Molecular Biology at Memorial Sloan–Kettering Cancer Center in New York...
and Nancy HopkinsNancy Hopkins (scientist)Nancy Hopkins, an American molecular biologist, is the Amgen, Inc. Professor of Biology at the Massachusetts Institute of Technology. She is a member of the National Academy of Sciences, the Institute of Medicine of the National Academy, and the American Academy of Arts and Sciences...
, "The Operators Controlled by the Lambda Phage Repressor", PNAS, v.60, n.4, pp.1282–1287 (1968). - Barbara J. Meyer, Dennis G. Kleid, and Mark Ptashne, "Lambda Repressor Turns Off Transcription of Its Own Gene", PNAS, v.72, n.12, pp.4785–4789 (December 1975).
- Gottesman, M. and Weisberg, R.A. 2004 "Little lambda - who made thee?", Micro and Mol Biol Revs, 68, 796-813 (available online at Microbiology and Molecular Biology Reviews, American Society for MicrobiologyAmerican Society for MicrobiologyThe American Society for Microbiology is a professional organization for scientists who study viruses, bacteria, fungi, algae, and protozoa as well as other aspects of microbiology. Microbiology is the study of organisms too small to be seen with the naked eye and which must be viewed with a...
) - Ptashne, M. "A Genetic Switch: Phage Lambda Revisited", 3rd edition 2003
- Snyder, L. and Champness, W. "Molecular Genetics of Bacteria", 3rd edition 2007 (Contains an informative and well illustrated overview of bacteriophage lambda)
- Splasho, Online overview of lambda (illustrates genes active at all stages in lifecycle)

