
Glutamate racemase
Encyclopedia
In enzymology, glutamate racemase (MurI with a capital i) is an enzyme
that catalyzes
the chemical reaction
Hence, this enzyme RacE has one substrate
, L-glutamate
, and one product
, D-glutamate.
This enzyme belongs to the family of isomerase
s, specifically those racemases and epimerases acting on amino acid
s and derivatives, including proline racemase, aspartate racemase, and diaminopimelate epimerase. This enzyme participates in glutamate metabolism that is essential for cell wall
biosynthesis
in bacteria. Glutamate racemase performs the additional function of gyrase inhibition, preventing gyrase from binding to DNA.
Glutamate racemase (MurI) serves two distinct metabolic functions: primarily, it is a critical enzyme in cell wall biosynthesis, but also plays a role in gyrase inhibition. The ability of glutamate racemase and other proteins to serve two distinct functions is known as "moonlighting
".
, it was generally believed by scientists that an enzyme
only had one function which lead to the concept of “one gene, one enzyme”. However, this concept no longer applies in science after the discovery that some proteins consist of both major and minor functions. This led to numerous studies attempting to relate the two functions to each other. The minor functions of these unique enzymes are called moonlighting functions, in which a protein can have a secondary functions not dependent upon the main function. These two functions of the moonlighting protein are found in a single polypeptide chain. Proteins that are multifunctional are not included due to gene fusion, families of homologous proteins, splice variants or promiscuous enzyme activities. The enzyme glutamate racemase (MurI) is an example of a moonlighting protein, functioning both in bacterial cell wall
biosynthesis as well as in gyrase inhibition.
compared to four-stranded β-sheets of C-domain. These structural specifications are not identical between MurI of different species; S. pyogenes
and B. subtilis
actually possess the most structurally similar MurI enzymes found as of now. It is also not rare to find MurI as a dimer
.
The active site, as it is evenly between the N-domain and C-domain, is also between the two cysteine
residues. It is accessible to solvents, as several water molecules, such as W1, are found in the active site. In some species, the active site also incorporates sulfate
ions to undergo hydrogen bonding on the amide backbone
and the side chain
s.
. This enzyme is most commonly known as being responsible for the synthesis of bacterial cell walls. Through experimentation it was found that this enzyme is able to construct these cell walls by synthesizing D-glutamate from L-glutamate through racemization
. D-glutamate is a monomer of the peptidoglycan
layer in prokaryotic cell walls. Peptidoglycan is an essential structural component of the bacterial cell wall. The peptidoglycan layer is also responsible for the rigidity of the cell wall. This process, in which MurI helps catalyze the interconversion of glutamate enantiomers, like L-Glutamate, into the essential D-glutamate, is also cofactor independent. As such it can proceed without needing an additional source, which would bind to an allosteric site, altering the enzyme shape to assist in catalyzing the reaction. Murl involves a two-step process to catalyze the glutamate enantiomers to D-glutamate. The first step is a deprotonation of the substrate to form an anion. Subsequently the substrate gets reprotaned. Once the glutamate is in the active site of the enzyme it undergoes a very large conformational change of its domains. This change helps superimpose the two catalytic cysteine residues, Cys73 and Cys184, located on either sides of the substrate at equal positions. Those domains mentioned earlier are symmetric and this symmetry suggests that this racemase activity of the protein may have evolved from gene duplication. Due to this main function of biosynthesis of bacterial cell walls MurI has been targeted as an antibacterial in drug discovery.
This function of MurI was discovered experimentally. DNA gyrase was incubated with the MurI enzyme and then added to a sample of DNA; the results of this experiment showed inhibition of supercoiling activity when MurI was present. The cell wall biosynthesis function of MurI is not directly related to its moonlighting function. MurI’s ability to inhibit gyrase binding can proceed independently of its main function. This means that DNA gyrase, in turn, will not have any effect on MurI's racemization, which was confirmed in a study of the racemization with and without the presence of DNA gyrase. In an experimental analysis, it was determined that MurI employs the use of two different enzymatic active sites for its two functions. This was shown by the inclusion of the racemase substrate L-glutamate in an assay with the separated gyrase inhibition site. The gyrase inhibition occurs in both supercoiling and relaxing activities of the DNA gyrase, and the study concluded that the inhibition activity was able to proceed, unchanged, in the presence of the racemase substrate. This dictates that the two functions can be carried out independently of each other, on non-overlapping sites, making MurI a true moonlighting protein. Mutant forms of MurI that are unable to exhibit their racemase function, no matter how compromised their racemase abilities were, were still proven through a study to be able to perform the DNA gyrase inhibition, with comparable results to a non-mutated form of MurI.
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
that catalyzes
Catalysis
Catalysis is the change in rate of a chemical reaction due to the participation of a substance called a catalyst. Unlike other reagents that participate in the chemical reaction, a catalyst is not consumed by the reaction itself. A catalyst may participate in multiple chemical transformations....
the chemical reaction
Chemical reaction
A chemical reaction is a process that leads to the transformation of one set of chemical substances to another. Chemical reactions can be either spontaneous, requiring no input of energy, or non-spontaneous, typically following the input of some type of energy, such as heat, light or electricity...
- L-glutamate
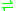 D-glutamate
D-glutamate
Hence, this enzyme RacE has one substrate
Substrate (biochemistry)
In biochemistry, a substrate is a molecule upon which an enzyme acts. Enzymes catalyze chemical reactions involving the substrate. In the case of a single substrate, the substrate binds with the enzyme active site, and an enzyme-substrate complex is formed. The substrate is transformed into one or...
, L-glutamate
Glutamic acid
Glutamic acid is one of the 20 proteinogenic amino acids, and its codons are GAA and GAG. It is a non-essential amino acid. The carboxylate anions and salts of glutamic acid are known as glutamates...
, and one product
Product (chemistry)
Product are formed during chemical reactions as reagents are consumed. Products have lower energy than the reagents and are produced during the reaction according to the second law of thermodynamics. The released energy comes from changes in chemical bonds between atoms in reagent molecules and...
, D-glutamate.
This enzyme belongs to the family of isomerase
Isomerase
In biochemistry, an isomerase is an enzyme that catalyzes the structural rearrangement of isomers. Isomerases thus catalyze reactions of the formwhere B is an isomer of A.-Nomenclature:...
s, specifically those racemases and epimerases acting on amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
s and derivatives, including proline racemase, aspartate racemase, and diaminopimelate epimerase. This enzyme participates in glutamate metabolism that is essential for cell wall
Cell wall
The cell wall is the tough, usually flexible but sometimes fairly rigid layer that surrounds some types of cells. It is located outside the cell membrane and provides these cells with structural support and protection, and also acts as a filtering mechanism. A major function of the cell wall is to...
biosynthesis
Biosynthesis
Biosynthesis is an enzyme-catalyzed process in cells of living organisms by which substrates are converted to more complex products. The biosynthesis process often consists of several enzymatic steps in which the product of one step is used as substrate in the following step...
in bacteria. Glutamate racemase performs the additional function of gyrase inhibition, preventing gyrase from binding to DNA.
Glutamate racemase (MurI) serves two distinct metabolic functions: primarily, it is a critical enzyme in cell wall biosynthesis, but also plays a role in gyrase inhibition. The ability of glutamate racemase and other proteins to serve two distinct functions is known as "moonlighting
Protein moonlighting
Protein moonlighting is a phenomenon by which a protein can perform more than one function. Ancestral moonlighting proteins originally possessed a single function but through evolution, acquired additional functions. Many proteins that moonlight are enzymes; others are receptors, ion channels or...
".
Moonlighting background
Before the discovery of moonlighting proteinsProtein moonlighting
Protein moonlighting is a phenomenon by which a protein can perform more than one function. Ancestral moonlighting proteins originally possessed a single function but through evolution, acquired additional functions. Many proteins that moonlight are enzymes; others are receptors, ion channels or...
, it was generally believed by scientists that an enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
only had one function which lead to the concept of “one gene, one enzyme”. However, this concept no longer applies in science after the discovery that some proteins consist of both major and minor functions. This led to numerous studies attempting to relate the two functions to each other. The minor functions of these unique enzymes are called moonlighting functions, in which a protein can have a secondary functions not dependent upon the main function. These two functions of the moonlighting protein are found in a single polypeptide chain. Proteins that are multifunctional are not included due to gene fusion, families of homologous proteins, splice variants or promiscuous enzyme activities. The enzyme glutamate racemase (MurI) is an example of a moonlighting protein, functioning both in bacterial cell wall
Cell wall
The cell wall is the tough, usually flexible but sometimes fairly rigid layer that surrounds some types of cells. It is located outside the cell membrane and provides these cells with structural support and protection, and also acts as a filtering mechanism. A major function of the cell wall is to...
biosynthesis as well as in gyrase inhibition.
Structure
The dimensions of MurI is approximately 35 Å × 40 Å × 45 Å and consists of two compact domains of α/β structure. With the active site in between the two domains, the N-terminal domain contains residues 1-97 and 207-264 while the C-terminal domain includes residues 98-206. This allows the enzyme to produce L-isomer from D-glutamate. Also, the N-domain is composed of five-stranded β-sheetsBeta sheet
The β sheet is the second form of regular secondary structure in proteins, only somewhat less common than the alpha helix. Beta sheets consist of beta strands connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet...
compared to four-stranded β-sheets of C-domain. These structural specifications are not identical between MurI of different species; S. pyogenes
Streptococcus pyogenes
Streptococcus pyogenes is a spherical, Gram-positive bacterium that is the cause of group A streptococcal infections. S. pyogenes displays streptococcal group A antigen on its cell wall. S...
and B. subtilis
Bacillus subtilis
Bacillus subtilis, known also as the hay bacillus or grass bacillus, is a Gram-positive, catalase-positive bacterium commonly found in soil. A member of the genus Bacillus, B. subtilis is rod-shaped, and has the ability to form a tough, protective endospore, allowing the organism to tolerate...
actually possess the most structurally similar MurI enzymes found as of now. It is also not rare to find MurI as a dimer
Protein dimer
In biochemistry, a dimer is a macromolecular complex formed by two, usually non-covalently bound, macromolecules like proteins or nucleic acids...
.
The active site, as it is evenly between the N-domain and C-domain, is also between the two cysteine
Cysteine
Cysteine is an α-amino acid with the chemical formula HO2CCHCH2SH. It is a non-essential amino acid, which means that it is biosynthesized in humans. Its codons are UGU and UGC. The side chain on cysteine is thiol, which is polar and thus cysteine is usually classified as a hydrophilic amino acid...
residues. It is accessible to solvents, as several water molecules, such as W1, are found in the active site. In some species, the active site also incorporates sulfate
Sulfate
In inorganic chemistry, a sulfate is a salt of sulfuric acid.-Chemical properties:...
ions to undergo hydrogen bonding on the amide backbone
Peptide bond
This article is about the peptide link found within biological molecules, such as proteins. A similar article for synthetic molecules is being created...
and the side chain
Side chain
In organic chemistry and biochemistry, a side chain is a chemical group that is attached to a core part of the molecule called "main chain" or backbone. The placeholder R is often used as a generic placeholder for alkyl group side chains in chemical structure diagrams. To indicate other non-carbon...
s.
Bacterial wall synthesis
Glutamate racemase is a bacterial enzyme that is encoded by the murI geneGene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
. This enzyme is most commonly known as being responsible for the synthesis of bacterial cell walls. Through experimentation it was found that this enzyme is able to construct these cell walls by synthesizing D-glutamate from L-glutamate through racemization
Racemization
In chemistry, racemization refers to the converting of an enantiomerically pure mixture into a mixture where more than one of the enantiomers are present...
. D-glutamate is a monomer of the peptidoglycan
Peptidoglycan
Peptidoglycan, also known as murein, is a polymer consisting of sugars and amino acids that forms a mesh-like layer outside the plasma membrane of bacteria , forming the cell wall. The sugar component consists of alternating residues of β- linked N-acetylglucosamine and N-acetylmuramic acid...
layer in prokaryotic cell walls. Peptidoglycan is an essential structural component of the bacterial cell wall. The peptidoglycan layer is also responsible for the rigidity of the cell wall. This process, in which MurI helps catalyze the interconversion of glutamate enantiomers, like L-Glutamate, into the essential D-glutamate, is also cofactor independent. As such it can proceed without needing an additional source, which would bind to an allosteric site, altering the enzyme shape to assist in catalyzing the reaction. Murl involves a two-step process to catalyze the glutamate enantiomers to D-glutamate. The first step is a deprotonation of the substrate to form an anion. Subsequently the substrate gets reprotaned. Once the glutamate is in the active site of the enzyme it undergoes a very large conformational change of its domains. This change helps superimpose the two catalytic cysteine residues, Cys73 and Cys184, located on either sides of the substrate at equal positions. Those domains mentioned earlier are symmetric and this symmetry suggests that this racemase activity of the protein may have evolved from gene duplication. Due to this main function of biosynthesis of bacterial cell walls MurI has been targeted as an antibacterial in drug discovery.
Gyrase inhibition
Along with its main function of cell wall biosynthesis, the moonlighting protein glutamate racemase also functions independently as a gyrase inhibitor. Present in certain forms of bacteria, MurI reduces the activity of DNA gyrase by preventing gyrase from binding to DNA. When gyrase binds to DNA, the enzyme decreases the tension in the DNA strands as they are unwound and causes the strands to become supercoiled. This is a critical step in DNA replication in these cells which results in the reproduction of bacterial cells. The presence of glutamate racemase in the process inhibits gyrase from effectively binding to DNA by deforming the shape of the enzyme’s active site. It essentially disallows gyrase from catalyzing the reaction that coils unwinding DNA strands.This function of MurI was discovered experimentally. DNA gyrase was incubated with the MurI enzyme and then added to a sample of DNA; the results of this experiment showed inhibition of supercoiling activity when MurI was present. The cell wall biosynthesis function of MurI is not directly related to its moonlighting function. MurI’s ability to inhibit gyrase binding can proceed independently of its main function. This means that DNA gyrase, in turn, will not have any effect on MurI's racemization, which was confirmed in a study of the racemization with and without the presence of DNA gyrase. In an experimental analysis, it was determined that MurI employs the use of two different enzymatic active sites for its two functions. This was shown by the inclusion of the racemase substrate L-glutamate in an assay with the separated gyrase inhibition site. The gyrase inhibition occurs in both supercoiling and relaxing activities of the DNA gyrase, and the study concluded that the inhibition activity was able to proceed, unchanged, in the presence of the racemase substrate. This dictates that the two functions can be carried out independently of each other, on non-overlapping sites, making MurI a true moonlighting protein. Mutant forms of MurI that are unable to exhibit their racemase function, no matter how compromised their racemase abilities were, were still proven through a study to be able to perform the DNA gyrase inhibition, with comparable results to a non-mutated form of MurI.

