
AP endonucleases
Encyclopedia
Endonuclease
Endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain, in contrast to exonucleases, which cleave phosphodiester bonds at the end of a polynucleotide chain. Typically, a restriction site will be a palindromic sequence four to six nucleotides long. Most...
(BRENDA
BRENDA
BRENDA is an enzyme information system representing one of the most comprehensive enzyme repositories.- Introduction :...
= 4.2.99.18) is an enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
that is involved in the DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
base excision repair
Base excision repair
In biochemistry and genetics, base excision repair is a cellular mechanism that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky...
pathway (BER). Its main role in the repair of damaged or mismatched nucleotides in DNA is to create a nick in the phosphodiester backbone of the AP site
AP site
In biochemistry and molecular genetics, an AP site , also known as an abasic site, is a location in DNA that has neither a purine nor a pyrimidine base, either spontaneously or due to DNA damage...
created when DNA glycosylase removes the damaged base. There are four types of AP endonucleases that have been classified according to their sites of incision. Class I and class II AP endonucleases incise DNA at the phosphate groups 3´ and 5´ to the baseless site leaving 3´-OH and 5´-phosphate termini. Class III and class IV AP endonucleases also cleave DNA at the phosphate groups 3´ and 5´to the baseless site, but they generate a 3´-phosphate and a 5´-OH. Human AP Endonuclease (APE1), like most AP endoucleases, is of class II and requires an Mg2+ in its active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
in order to carry out its role in base excision repair.
Structure of APE1
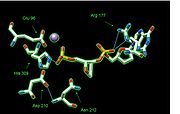
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
residues that enable it to react selectively with AP sites. Three APE1 residues (Arg
Arginine
Arginine is an α-amino acid. The L-form is one of the 20 most common natural amino acids. At the level of molecular genetics, in the structure of the messenger ribonucleic acid mRNA, CGU, CGC, CGA, CGG, AGA, and AGG, are the triplets of nucleotide bases or codons that codify for arginine during...
73, Ala
Alanine
Alanine is an α-amino acid with the chemical formula CH3CHCOOH. The L-isomer is one of the 20 amino acids encoded by the genetic code. Its codons are GCU, GCC, GCA, and GCG. It is classified as a nonpolar amino acid...
74, and Lys
Lysine
Lysine is an α-amino acid with the chemical formula HO2CCH4NH2. It is an essential amino acid, which means that the human body cannot synthesize it. Its codons are AAA and AAG....
78) contact three consecutive DNA phosphates on the strand opposite the one containing the AP site while Tyr
Tyrosine
Tyrosine or 4-hydroxyphenylalanine, is one of the 22 amino acids that are used by cells to synthesize proteins. Its codons are UAC and UAU. It is a non-essential amino acid with a polar side group...
128 and Gly
Glycine
Glycine is an organic compound with the formula NH2CH2COOH. Having a hydrogen substituent as its 'side chain', glycine is the smallest of the 20 amino acids commonly found in proteins. Its codons are GGU, GGC, GGA, GGG cf. the genetic code.Glycine is a colourless, sweet-tasting crystalline solid...
127 span and widen the minor groove, anchoring the DNA for the extreme kinking caused by the interaction between positive residues found in four loops and one α-helix and the negative phosphate groups found in the phosphodiester backbone of DNA.
This extreme kinking forces the baseless portion of DNA into APE1’s active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
. This active site is bordered by Phe
Phenylalanine
Phenylalanine is an α-amino acid with the formula C6H5CH2CHCOOH. This essential amino acid is classified as nonpolar because of the hydrophobic nature of the benzyl side chain. L-Phenylalanine is an electrically neutral amino acid, one of the twenty common amino acids used to biochemically form...
266, Trp
Tryptophan
Tryptophan is one of the 20 standard amino acids, as well as an essential amino acid in the human diet. It is encoded in the standard genetic code as the codon UGG...
280, and Leu
Leucine
Leucine is a branched-chain α-amino acid with the chemical formula HO2CCHCH2CH2. Leucine is classified as a hydrophobic amino acid due to its aliphatic isobutyl side chain. It is encoded by six codons and is a major component of the subunits in ferritin, astacin and other 'buffer' proteins...
282, which pack tightly with the hydrophobic side of the AP site, discriminating against sites that do have bases. The AP site is then further stabilized through hydrogen bonding of the phosphate group 5´ to the AP site with Asn
Asparagine
Asparagine is one of the 20 most common natural amino acids on Earth. It has carboxamide as the side-chain's functional group. It is not an essential amino acid...
174, Asn212, His
Histidine
Histidine Histidine, an essential amino acid, has a positively charged imidazole functional group. It is one of the 22 proteinogenic amino acids. Its codons are CAU and CAC. Histidine was first isolated by German physician Albrecht Kossel in 1896. Histidine is an essential amino acid in humans...
309, and the Mg2+ ion while its orphan base partner is stabilized through hydrogen bonding with Met
Methionine
Methionine is an α-amino acid with the chemical formula HO2CCHCH2CH2SCH3. This essential amino acid is classified as nonpolar. This amino-acid is coded by the codon AUG, also known as the initiation codon, since it indicates mRNA's coding region where translation into protein...
270. The phosphate group 3' to the AP site is stabilized through hydrogen bonding to Arg
Arginine
Arginine is an α-amino acid. The L-form is one of the 20 most common natural amino acids. At the level of molecular genetics, in the structure of the messenger ribonucleic acid mRNA, CGU, CGC, CGA, CGG, AGA, and AGG, are the triplets of nucleotide bases or codons that codify for arginine during...
177. Meanwhile, an Asp
Aspartic acid
Aspartic acid is an α-amino acid with the chemical formula HOOCCHCH2COOH. The carboxylate anion, salt, or ester of aspartic acid is known as aspartate. The L-isomer of aspartate is one of the 20 proteinogenic amino acids, i.e., the building blocks of proteins...
210 in the active site, which is made more reactive due to the increase in its pKa (or the negative log of acid dissociation constant
Acid dissociation constant
An acid dissociation constant, Ka, is a quantitative measure of the strength of an acid in solution. It is the equilibrium constant for a chemical reaction known as dissociation in the context of acid-base reactions...
) caused through its stabilization through its hydrogen bonding between Asn68 and Asn212, activates the nucleophile
Nucleophile
A nucleophile is a species that donates an electron-pair to an electrophile to form a chemical bond in a reaction. All molecules or ions with a free pair of electrons can act as nucleophiles. Because nucleophiles donate electrons, they are by definition Lewis bases.Nucleophilic describes the...
that attacks and cleaves the phosphodiester backbone and probably results in the observed maximal APE1 activity at a pH
PH
In chemistry, pH is a measure of the acidity or basicity of an aqueous solution. Pure water is said to be neutral, with a pH close to 7.0 at . Solutions with a pH less than 7 are said to be acidic and solutions with a pH greater than 7 are basic or alkaline...
of 7.5.
Mechanism
The APE1 enzyme creates a nick in the phosphodiester backbone at a abasic(baseless) site through a simple acyl substitution mechanism. First, the Asp210 residue in the active site deprotonates a water molecule, which can then perform a nucleophilic attack on the phosphate group located 5´ to the AP site. Next, electrons from one of the oxygen atom in the phosphate group moves down, kicking off one of the other oxygen to create a free 5´ phosphate group on the AP site and a free 3´-OH on the normal nucleotide, both of which are stabilized by the Mg2+ ion.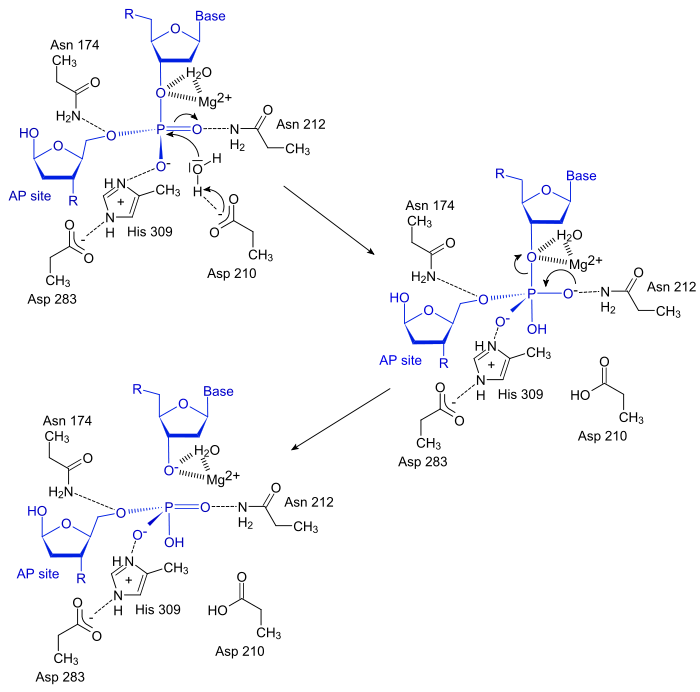
Inhibition of APE1

Lucanthone
Lucanthone is a drug used in chemotherapy.Lucanthone is a prodrug and is converted to an active metabolite "hycanthone".-Mechanism of action:...
. Both of these structures possess rings attached to short chains, which appear similar to the deoxyribose sugar ring without a base attached and phosphodiester bond in DNA. Both also contain lots of H-bond acceptors which may interact with the H-bond donors in the active site of APE1, causing these inhibitors to stick in the active site and preventing the enzyme from catalyzing other reactions.
APE1 as Chemopreventive Target
Because APE1 performs an essential function in DNA base-excision repair pathway, it has become a target for researchers looking for means to prevent cancer cells from surviving chemotherapy. Not only is APE1 needed in and of itself to create the nick in the DNA backbone so that the enzymes involved later in the BER pathway can recognize the AP-site, it also has a redox function that helps activate other enzymes involved in DNA repair. As such, knocking down APE1 could lead to tumor cell sensitivity, thus preventing cancer cells from persisting after chemotherapy.External links
- Basic Definition
- AP endonucleases family 1 in PROSITEPROSITEPROSITE is a protein database. It consists of entries describing the protein families, domains and functional sites as well as amino acid patterns, signatures, and profiles in them. These are manually curated by a team of the Swiss Institute of Bioinformatics and tightly integrated into Swiss-Prot...
- AP endonucleases family 2 in PROSITEPROSITEPROSITE is a protein database. It consists of entries describing the protein families, domains and functional sites as well as amino acid patterns, signatures, and profiles in them. These are manually curated by a team of the Swiss Institute of Bioinformatics and tightly integrated into Swiss-Prot...
- Application in Long Patch Base Excision Repair
- Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells

