
SmY RNA
Encyclopedia
SmY ribonucleic acids are a family of small nuclear RNA
s found in some species of nematode
worms. They are thought to be involved in mRNA trans-splicing
.
SmY RNAs are about 70–90 nucleotide
s long and share a common secondary structure
, with two stem-loop
s flanking a consensus binding site
for Sm protein
. Sm protein is a shared component of spliceosomal
snRNP
s.
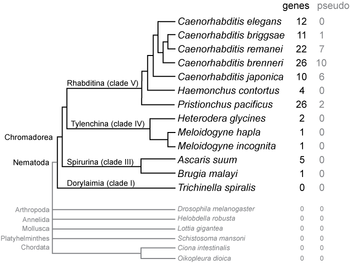 SmY RNAs have been found in nematodes of class Chromadorea, which includes the most commonly studied nematodes (such as Caenorhabditis
SmY RNAs have been found in nematodes of class Chromadorea, which includes the most commonly studied nematodes (such as Caenorhabditis
, Pristionchus, and Ascaris
), but not in the more distantly related Trichinella spiralis
in class Dorylaimia. The number of SmY genes in each species varies, with most Caenorhabditis and Pristionchus species having 10–26 related paralogous copies, while other nematodes have 1–5.
spliceosome
preparations, as was another called SmX RNA that is not detectably homologous
to SmY. Twelve SmY homologs were identified computationally
in Caenorhabditis elegans
, and ten in Caenorhabditis briggsae
. Several transcripts
from these SmY gene
s were cloned
and sequenced
in a systematic survey of small non-coding RNA
transcripts in C. elegans.
A systematic survey of 2,2,7-trimethylguanosine (TMG) 5' capped
transcripts in C.elegans using anti TMG antibodies identified two TMG capped SmY transcripts. Sequence analysis of the potential Sm binding sites in these transcripts indicated the SmY, U5 snRNA
, U3 snoRNA
and the spliced leader RNAs transcripts (SL1 and SL2
) all contain a very similar consensus SM binding sequence ( AAU4–5GGA). The predicted SM binding sites identified in the U1
, U2
and U4
snRNA transcripts varied from this consensus.
, SmY RNAs copurify
with spliceosome
and with Sm, SL75p, and SL26p proteins, while the better-characterized C. elegans SL1
trans-splicing snRNA copurifies in a complex with Sm, SL75p, and SL21p (a paralog of SL26p). Loss of function of either SL21p or SL26p individually causes only a weak cold-sensitive phenotype
, whereas knockdown of both is lethal, as is a SL75p knockdown. Based on these results, the SmY RNAs are believed to have a function in trans-splicing.
Small nuclear RNA
Small nuclear ribonucleic acid is a class of small RNA molecules that are found within the nucleus of eukaryotic cells. They are transcribed by RNA polymerase II or RNA polymerase III and are involved in a variety of important processes such as RNA splicing , regulation of transcription factors ...
s found in some species of nematode
Nematode
The nematodes or roundworms are the most diverse phylum of pseudocoelomates, and one of the most diverse of all animals. Nematode species are very difficult to distinguish; over 28,000 have been described, of which over 16,000 are parasitic. It has been estimated that the total number of nematode...
worms. They are thought to be involved in mRNA trans-splicing
Trans-splicing
Trans-splicing is a special form of RNA processing in eukaryotes where exons from two different primary RNA transcripts are joined end to end and ligated....
.
SmY RNAs are about 70–90 nucleotide
Nucleotide
Nucleotides are molecules that, when joined together, make up the structural units of RNA and DNA. In addition, nucleotides participate in cellular signaling , and are incorporated into important cofactors of enzymatic reactions...
s long and share a common secondary structure
Secondary structure
In biochemistry and structural biology, secondary structure is the general three-dimensional form of local segments of biopolymers such as proteins and nucleic acids...
, with two stem-loop
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions,...
s flanking a consensus binding site
Binding site
In biochemistry, a binding site is a region on a protein, DNA, or RNA to which specific other molecules and ions—in this context collectively called ligands—form a chemical bond...
for Sm protein
LSm
In biology, LSm proteins are a family of RNA-binding proteins found in virtually every cellular organism. LSm is a contraction of 'like Sm', because the first identified members of the LSm protein family were the Sm proteins...
. Sm protein is a shared component of spliceosomal
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
snRNP
SnRNP
snRNPs , or small nuclear ribonucleoproteins, are RNA-protein complexes that combine with unmodified pre-mRNA and various other proteins to form a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
s.

Caenorhabditis
Caenorhabditis is a genus of nematodes which live in bacteria-rich environments like compost piles and decaying dead animals. It contains the noted model organism Caenorhabditis elegans and several other species for which a genome sequence is either available or currently being determined. The two...
, Pristionchus, and Ascaris
Ascaris
Ascaris is a genus of parasitic nematode worms known as the "giant intestinal roundworms". One species, A. suum, typically infects pigs, while another, A. lumbricoides, affects human populations, typically in sub-tropical and tropical areas with poor sanitation. A...
), but not in the more distantly related Trichinella spiralis
Trichinella spiralis
Trichinella spiralis is a nematode parasite, occurring in rats, pigs, bears and humans, and is responsible for the disease trichinosis. It is sometimes referred to as the "pork worm" due to it being found commonly in undercooked pork products...
in class Dorylaimia. The number of SmY genes in each species varies, with most Caenorhabditis and Pristionchus species having 10–26 related paralogous copies, while other nematodes have 1–5.
Discovery
The first SmY RNA was discovered in 1996 in purified Ascaris lumbricoidesAscaris
Ascaris is a genus of parasitic nematode worms known as the "giant intestinal roundworms". One species, A. suum, typically infects pigs, while another, A. lumbricoides, affects human populations, typically in sub-tropical and tropical areas with poor sanitation. A...
spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
preparations, as was another called SmX RNA that is not detectably homologous
Homology (biology)
Homology forms the basis of organization for comparative biology. In 1843, Richard Owen defined homology as "the same organ in different animals under every variety of form and function". Organs as different as a bat's wing, a seal's flipper, a cat's paw and a human hand have a common underlying...
to SmY. Twelve SmY homologs were identified computationally
Computational biology
Computational biology involves the development and application of data-analytical and theoretical methods, mathematical modeling and computational simulation techniques to the study of biological, behavioral, and social systems...
in Caenorhabditis elegans
Caenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode , about 1 mm in length, which lives in temperate soil environments. Research into the molecular and developmental biology of C. elegans was begun in 1974 by Sydney Brenner and it has since been used extensively as a model...
, and ten in Caenorhabditis briggsae
Caenorhabditis briggsae
Caenorhabditis briggsae is a small nematode, closely related to Caenorhabditis elegans. The differences between the two species are subtle. The male tail in C. briggsae has a slightly different morphology than C. elegans. Other differences include changes in vulval precursor competence and the...
. Several transcripts
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
from these SmY gene
Gene
A gene is a molecular unit of heredity of a living organism. It is a name given to some stretches of DNA and RNA that code for a type of protein or for an RNA chain that has a function in the organism. Living beings depend on genes, as they specify all proteins and functional RNA chains...
s were cloned
Molecular cloning
Molecular cloning refers to a set of experimental methods in molecular biology that are used to assemble recombinant DNA molecules and to direct their replication within host organisms...
and sequenced
Sequencing
In genetics and biochemistry, sequencing means to determine the primary structure of an unbranched biopolymer...
in a systematic survey of small non-coding RNA
Non-coding RNA
A non-coding RNA is a functional RNA molecule that is not translated into a protein. Less-frequently used synonyms are non-protein-coding RNA , non-messenger RNA and functional RNA . The term small RNA is often used for short bacterial ncRNAs...
transcripts in C. elegans.
A systematic survey of 2,2,7-trimethylguanosine (TMG) 5' capped
5' cap
The 5' cap is a specially altered nucleotide on the 5' end of precursor messenger RNA and some other primary RNA transcripts as found in eukaryotes. The process of 5' capping is vital to creating mature messenger RNA, which is then able to undergo translation...
transcripts in C.elegans using anti TMG antibodies identified two TMG capped SmY transcripts. Sequence analysis of the potential Sm binding sites in these transcripts indicated the SmY, U5 snRNA
U5 spliceosomal RNA
U5 RNA is a non-coding RNA that is a component of both types of known spliceosome. The precise function of this molecule is unknown, though it is known that the 5' loop is required for splice site selection and p220 binding, and that both the 3' stem-loop and the Sm site are important for Sm...
, U3 snoRNA
Small nucleolar RNA U3
U3 snoRNA is a non-coding RNA found predominantly in the nucleolus.U3 has C/D box motifs that technically make it a member of the box C/D class of snoRNAs; however, unlike other C/D box snoRNAs, it has not been shown to direct 2'-O-methylation of other RNAs.Rather, U3 is thought to guide...
and the spliced leader RNAs transcripts (SL1 and SL2
SL2
SL2 was the name of a hardcore/rave music act in the early 1990s from London, England. They also recorded, remixed or produced under the names Slipmatt & Lime and T.H.C.-Background:...
) all contain a very similar consensus SM binding sequence ( AAU4–5GGA). The predicted SM binding sites identified in the U1
U1 spliceosomal RNA
U1 spliceosomal RNA is the small nuclear RNA component of U1 snRNP , an RNA-protein complex that combines with other snRNPs, unmodified pre-mRNA, and various other proteins to assemble a spliceosome, a large RNA-protein molecular complex upon which splicing of pre-mRNA occurs...
, U2
U2 spliceosomal RNA
U2 spliceosomal RNA is a small nuclear RNA component of the spliceosome . Complementary binding between U2 snRNA and the branchpoint sequence of the intron results in the bulging out of an unpaired adenosine, on the BPS, which initiates a nucleophilic attack at the intronic 5' splice...
and U4
U4 spliceosomal RNA
The U4 small nuclear Ribo-Nucleic Acid is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA...
snRNA transcripts varied from this consensus.
Function
In C. elegansCaenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode , about 1 mm in length, which lives in temperate soil environments. Research into the molecular and developmental biology of C. elegans was begun in 1974 by Sydney Brenner and it has since been used extensively as a model...
, SmY RNAs copurify
Copurification
Copurification in a chemical or biochemical context is the physical separation by chromatography or other purification technique of two or more substances of interest from other contaminating substances...
with spliceosome
Spliceosome
A spliceosome is a complex of snRNA and protein subunits that removes introns from a transcribed pre-mRNA segment. This process is generally referred to as splicing.-Composition:...
and with Sm, SL75p, and SL26p proteins, while the better-characterized C. elegans SL1
SL1 RNA
This family represents the SL1 RNA. SL1 is commonly located in the spacer region between 5S-rRNA genes where it is involved in trans-splicing. Trans-splicing is a form of RNA processing. The acquisition of a spliced leader from an SL RNA is an intra-molecular reaction which precisely joins exons...
trans-splicing snRNA copurifies in a complex with Sm, SL75p, and SL21p (a paralog of SL26p). Loss of function of either SL21p or SL26p individually causes only a weak cold-sensitive phenotype
Phenotype
A phenotype is an organism's observable characteristics or traits: such as its morphology, development, biochemical or physiological properties, behavior, and products of behavior...
, whereas knockdown of both is lethal, as is a SL75p knockdown. Based on these results, the SmY RNAs are believed to have a function in trans-splicing.

