
Phosphoenolpyruvate mutase
Encyclopedia
In enzymology, a phosphoenolpyruvate mutase is an enzyme
that catalyzes
the chemical reaction
Hence, this enzyme has one substrate
, phosphoenolpyruvate
(PEP), and one product
, 3-phosphonopyruvate (PPR), which are structural isomers.
This enzyme belongs to the family of isomerase
s, specifically the phosphotransferases (phosphomutases), which transfer phosphate groups within a molecule. The systematic name of this enzyme class is phosphoenolpyruvate 2,3-phosphonomutase. Other names in common use include phosphoenolpyruvate-phosphonopyruvate phosphomutase, PEP phosphomutase, phosphoenolpyruvate phosphomutase, PEPPM, and PEP phosphomutase. This enzyme participates in aminophosphonate metabolism.
Phosphoenolpyruvate mutase was discovered in 1988.
have been solved for this class of enzymes, all by the Herzberg group http://carb.umbi.umd.edu/herzberg/research at the University of Maryland
using PEPPM from the blue mussel
, Mytilus edulis. The first structure (PDB
accession code ) was solved in 1999 and featured a magnesium oxalate inhibitor . This structure identified the enzyme as consisting of identical beta barrel
subunits (exhibiting the TIM barrel
fold, which consists of eight parallel beta strands). Dimerization was observed in which a helix from each subunit interacts with the other subunit's barrel; the authors called this feature "helix swapping." The dimers can dimerize as well to form a homotetrameric enzyme. A double phosphoryl transfer mechanism was proposed on the basis of this study: this would involve breakage of PEP's phosphorus-oxygen bond to form a phosphoenzyme intermediate, followed by transfer of the phosphoryl group from the enzyme to carbon-3, forming PPR.
However, more recently, a structure with a sulfopyruvate inhibitor, which is a closer substrate analogue, was solved ; this study supported instead a dissociative mechanism. A notable feature of these structures was the shielding of the active site
from solvent; it was proposed that a significant conformational change
takes place on binding to allow this, moving the protein from an "open" to a "closed" state, and this was supported by several crystal structures in the open state . Three of these were of the wild type
: the apoenzyme in , the enzyme plus its magnesium ion cofactor in , and the enzyme at high ionic strength in . A mutant (D58A, in one of the active-site loops) was crystallized as an apoenzyme also . From these structures, an active-site "gating" loop (residues 115-133) that shields the substrate from solvent in the closed conformation was identified.
The two conformations, taken from the crystal structures 1M1B (closed) and 1S2T (open), are docked into each other in the images below; they differ negligibly except in the gating loop, which is colored purple for the closed conformation and blue for the open conformation. In the active-site closeup (left), several sidechains (cyan) that have been identified as important in catalysis are included as well; the overview (right) illustrates the distinctive helix-swapping fold. The images are still shots from ribbon
kinemage
s. Both of these structures were crystallized as dimers. In chain A (used for the active-site closeup), helices are red while loops (other than the gating loop) are white and beta strands are green; in chain B, helices are yellow, beta strands are olive, and loops are gray; these colors are the same for the closed and open structures. Magnesium ions are gray and the sulfopyruvate ligands are pink; both are from the closed structure (though the enzyme has also been crystallized with only magnesium bound, and it adopted an open conformation).
The structure of PEPPM is very similar to that of methylisocitrate lyase
, an enzyme involved in propanoate metabolism whose substrate is also a low-molecular weight carboxylic acid
--the beta-barrel structure as well as the active site layout and multimerization geometry are the same. Isocitrate lyase
is also quite similar, though each subunit has a second, smaller beta domain in addition to the main beta barrel.
Residues in the active-site gating loop, particularly Lys120, Asn122, and Leu124, also appear to interact with the substrate and reactive intermediate; these interactions explain why the loop moves into the closed conformation on substrate binding.
s, such as phosphonolipids and the antibiotics fosfomycin
and bialaphos. The formation of this bond is quite thermodynamically unfavorable; even though PEP is a very high-energy phosphate compound, the equilibrium in PEP-PPR interconversion still favors PEP . The enzyme phosphonopyruvate decarboxylase
presents a solution to this problem: it catalyzes the very thermodynamically favorable decarboxylation of PPR, and the resulting 2-phosphonoacetaldehyde is then converted into biologically useful phosphonates. This allows phosphoneolpyruvate's reaction to proceed in the forward direction, due to Le Chatelier's principle
. The decarboxylation removes product quickly, and thus the reaction moves forward even though there would be much more reactant than product if the system were allowed to reach equilibrium by itself.
The enzyme carboxyphosphoenolpyruvate phosphonomutase performs a similar reaction, converting P-carboxyphosphoenolpyruvate to phosphinopyruvate and carbon dioxide
. http://BioCyc.org/META/NEW-IMAGE?type=REACTION&object=2.7.8.23-RXN
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
that catalyzes
Catalysis
Catalysis is the change in rate of a chemical reaction due to the participation of a substance called a catalyst. Unlike other reagents that participate in the chemical reaction, a catalyst is not consumed by the reaction itself. A catalyst may participate in multiple chemical transformations....
the chemical reaction
Chemical reaction
A chemical reaction is a process that leads to the transformation of one set of chemical substances to another. Chemical reactions can be either spontaneous, requiring no input of energy, or non-spontaneous, typically following the input of some type of energy, such as heat, light or electricity...
- phosphoenolpyruvate
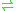 3-phosphonopyruvate
3-phosphonopyruvate
Hence, this enzyme has one substrate
Substrate (biochemistry)
In biochemistry, a substrate is a molecule upon which an enzyme acts. Enzymes catalyze chemical reactions involving the substrate. In the case of a single substrate, the substrate binds with the enzyme active site, and an enzyme-substrate complex is formed. The substrate is transformed into one or...
, phosphoenolpyruvate
Phosphoenolpyruvate
Phosphoenolpyruvic acid , or phosphoenolpyruvate as the anion, is an important chemical compound in biochemistry. It has the high-energy phosphate bond found in living organisms, and is involved in glycolysis and gluconeogenesis...
(PEP), and one product
Product (chemistry)
Product are formed during chemical reactions as reagents are consumed. Products have lower energy than the reagents and are produced during the reaction according to the second law of thermodynamics. The released energy comes from changes in chemical bonds between atoms in reagent molecules and...
, 3-phosphonopyruvate (PPR), which are structural isomers.
This enzyme belongs to the family of isomerase
Isomerase
In biochemistry, an isomerase is an enzyme that catalyzes the structural rearrangement of isomers. Isomerases thus catalyze reactions of the formwhere B is an isomer of A.-Nomenclature:...
s, specifically the phosphotransferases (phosphomutases), which transfer phosphate groups within a molecule. The systematic name of this enzyme class is phosphoenolpyruvate 2,3-phosphonomutase. Other names in common use include phosphoenolpyruvate-phosphonopyruvate phosphomutase, PEP phosphomutase, phosphoenolpyruvate phosphomutase, PEPPM, and PEP phosphomutase. This enzyme participates in aminophosphonate metabolism.
Phosphoenolpyruvate mutase was discovered in 1988.
Structural studies
As of late 2007, 6 structuresTertiary structure
In biochemistry and molecular biology, the tertiary structure of a protein or any other macromolecule is its three-dimensional structure, as defined by the atomic coordinates.-Relationship to primary structure:...
have been solved for this class of enzymes, all by the Herzberg group http://carb.umbi.umd.edu/herzberg/research at the University of Maryland
University of Maryland
When the term "University of Maryland" is used without any qualification, it generally refers to the University of Maryland, College Park.University of Maryland may refer to the following:...
using PEPPM from the blue mussel
Blue mussel
The blue mussel, Mytilus edulis, is a medium-sized edible marine bivalve mollusc in the family Mytilidae. In spite of its specific name edulis, it is not the sole edible Mytilus species.-Distribution:...
, Mytilus edulis. The first structure (PDB
Protein Data Bank
The Protein Data Bank is a repository for the 3-D structural data of large biological molecules, such as proteins and nucleic acids....
accession code ) was solved in 1999 and featured a magnesium oxalate inhibitor . This structure identified the enzyme as consisting of identical beta barrel
Beta barrel
A beta barrel is a large beta-sheet that twists and coils to form a closed structure in which the first strand is hydrogen bonded to the last.Beta-strands in beta-barrels are typically arranged in an antiparallel fashion...
subunits (exhibiting the TIM barrel
TIM barrel
The TIM barrel is a conserved protein fold consisting of eight α-helices and eight parallel β-strands that alternate along the peptide backbone. The structure is named after triosephosphate isomerase, a conserved glycolytic enzyme. TIM barrels are quite common among the conserved protein folds...
fold, which consists of eight parallel beta strands). Dimerization was observed in which a helix from each subunit interacts with the other subunit's barrel; the authors called this feature "helix swapping." The dimers can dimerize as well to form a homotetrameric enzyme. A double phosphoryl transfer mechanism was proposed on the basis of this study: this would involve breakage of PEP's phosphorus-oxygen bond to form a phosphoenzyme intermediate, followed by transfer of the phosphoryl group from the enzyme to carbon-3, forming PPR.
However, more recently, a structure with a sulfopyruvate inhibitor, which is a closer substrate analogue, was solved ; this study supported instead a dissociative mechanism. A notable feature of these structures was the shielding of the active site
Active site
In biology the active site is part of an enzyme where substrates bind and undergo a chemical reaction. The majority of enzymes are proteins but RNA enzymes called ribozymes also exist. The active site of an enzyme is usually found in a cleft or pocket that is lined by amino acid residues that...
from solvent; it was proposed that a significant conformational change
Conformational change
A macromolecule is usually flexible and dynamic. It can change its shape in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a conformational change...
takes place on binding to allow this, moving the protein from an "open" to a "closed" state, and this was supported by several crystal structures in the open state . Three of these were of the wild type
Wild type
Wild type refers to the phenotype of the typical form of a species as it occurs in nature. Originally, the wild type was conceptualized as a product of the standard, "normal" allele at a locus, in contrast to that produced by a non-standard, "mutant" allele...
: the apoenzyme in , the enzyme plus its magnesium ion cofactor in , and the enzyme at high ionic strength in . A mutant (D58A, in one of the active-site loops) was crystallized as an apoenzyme also . From these structures, an active-site "gating" loop (residues 115-133) that shields the substrate from solvent in the closed conformation was identified.
The two conformations, taken from the crystal structures 1M1B (closed) and 1S2T (open), are docked into each other in the images below; they differ negligibly except in the gating loop, which is colored purple for the closed conformation and blue for the open conformation. In the active-site closeup (left), several sidechains (cyan) that have been identified as important in catalysis are included as well; the overview (right) illustrates the distinctive helix-swapping fold. The images are still shots from ribbon
Ribbon diagram
Proteins are biological macromolecules made up of a long polypeptide chain of amino acids linked by peptide bonds...
kinemage
Kinemage
A kinemage is an interactive graphic scientific illustration. It often is used to visualize molecules, especially proteins and nucleic acids, although it can also represent other types of 3-dimensional data...
s. Both of these structures were crystallized as dimers. In chain A (used for the active-site closeup), helices are red while loops (other than the gating loop) are white and beta strands are green; in chain B, helices are yellow, beta strands are olive, and loops are gray; these colors are the same for the closed and open structures. Magnesium ions are gray and the sulfopyruvate ligands are pink; both are from the closed structure (though the enzyme has also been crystallized with only magnesium bound, and it adopted an open conformation).
The structure of PEPPM is very similar to that of methylisocitrate lyase
Methylisocitrate lyase
In enzymology, a methylisocitrate lyase is an enzyme that catalyzes the chemical reaction-3-hydroxybutane-1,2,3-tricarboxylate \rightleftharpoons pyruvate + succinate...
, an enzyme involved in propanoate metabolism whose substrate is also a low-molecular weight carboxylic acid
Carboxylic acid
Carboxylic acids are organic acids characterized by the presence of at least one carboxyl group. The general formula of a carboxylic acid is R-COOH, where R is some monovalent functional group...
--the beta-barrel structure as well as the active site layout and multimerization geometry are the same. Isocitrate lyase
Isocitrate lyase
Isocitrate lyase , or ICL, is an enzyme in the glyoxylate cycle that catalyzes the cleavage of isocitrate to succinate and glyoxylate. Together with malate synthase, it bypasses the two decarboxylation steps of the tricarboxylic acid cycle and is used by bacteria, fungi, and plants.The systematic...
is also quite similar, though each subunit has a second, smaller beta domain in addition to the main beta barrel.
Mechanism
Phosphoenolpyruvate mutase is thought to exhibit a dissociative mechanism. A magnesium ion is involved as a cofactor. The phosphoryl/phosphate group also appears to interact ionically with Arg159 and His190, stabilizing the reactive intermediate. A phosphoenzyme intermediate is unlikely because the most feasible residues for the covalent adduct can be mutated with only partial loss of function. The reaction involves dissociation of phosphorus from oxygen 2 and then a nucleophilic attack by carbon 3 on phosphorus. Notably, the configuration is retained at phosphorus, i.e. carbon 3 of PPR adds to the same face of phosphorus from which oxygen 2 of PEP was removed; this would be unlikely for a non-enzyme-catalyzed dissociative mechanism, but since the reactive intermediate interacts strongly with the amino acids and magnesium ions of the active site, it is to be expected in the presence of enzyme catalysis.Residues in the active-site gating loop, particularly Lys120, Asn122, and Leu124, also appear to interact with the substrate and reactive intermediate; these interactions explain why the loop moves into the closed conformation on substrate binding.
Biological function
Because phosphoenolpyruvate mutase has the unusual ability to form a new carbon-phosphorus bond, it is essential to the synthesis of phosphonatePhosphonate
Phosphonates or phosphonic acids are organic compounds containing C-PO2 or C-PO2 groups . Bisphosphonates were first synthesized in 1897 by Von Baeyer and Hofmann. An example of such a bisphosphonate is HEDP . Since the work of Schwarzenbach in 1949, phosphonic acids are known as effective...
s, such as phosphonolipids and the antibiotics fosfomycin
Fosfomycin
Fosfomycin is a broad-spectrum antibiotic produced by certain Streptomyces species.-Uses:...
and bialaphos. The formation of this bond is quite thermodynamically unfavorable; even though PEP is a very high-energy phosphate compound, the equilibrium in PEP-PPR interconversion still favors PEP . The enzyme phosphonopyruvate decarboxylase
Phosphonopyruvate decarboxylase
In enzymology, a phosphonopyruvate decarboxylase is an enzyme that catalyzes the chemical reactionHence, this enzyme has one substrate, 3-phosphonopyruvate, and two products, 2-phosphonoacetaldehyde and CO2....
presents a solution to this problem: it catalyzes the very thermodynamically favorable decarboxylation of PPR, and the resulting 2-phosphonoacetaldehyde is then converted into biologically useful phosphonates. This allows phosphoneolpyruvate's reaction to proceed in the forward direction, due to Le Chatelier's principle
Le Châtelier's principle
In chemistry, Le Chatelier's principle, also called the Chatelier's principle, can be used to predict the effect of a change in conditions on a chemical equilibrium. The principle is named after Henry Louis Le Chatelier and sometimes Karl Ferdinand Braun who discovered it independently...
. The decarboxylation removes product quickly, and thus the reaction moves forward even though there would be much more reactant than product if the system were allowed to reach equilibrium by itself.
The enzyme carboxyphosphoenolpyruvate phosphonomutase performs a similar reaction, converting P-carboxyphosphoenolpyruvate to phosphinopyruvate and carbon dioxide
Carbon dioxide
Carbon dioxide is a naturally occurring chemical compound composed of two oxygen atoms covalently bonded to a single carbon atom...
. http://BioCyc.org/META/NEW-IMAGE?type=REACTION&object=2.7.8.23-RXN

