Lysine 2,3-aminomutase
Encyclopedia
General Reaction
Lysine 2,3-aminomutase (KAM or LAM) (ECEC number
The Enzyme Commission number is a numerical classification scheme for enzymes, based on the chemical reactions they catalyze....
5.4.3.2) is an enzyme
Enzyme
Enzymes are proteins that catalyze chemical reactions. In enzymatic reactions, the molecules at the beginning of the process, called substrates, are converted into different molecules, called products. Almost all chemical reactions in a biological cell need enzymes in order to occur at rates...
which facilitates the conversion of the amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
lysine
Lysine
Lysine is an α-amino acid with the chemical formula HO2CCH4NH2. It is an essential amino acid, which means that the human body cannot synthesize it. Its codons are AAA and AAG....
to beta-lysine
Beta-lysine
Beta-lysine is an amino acid produced by platelets during coagulation and is directly antibacterial by causing lysis of many Gram positive bacteria by acting as a cationic detergent....
. It accomplishes this interconversion using three cofactors
Cofactor (biochemistry)
A cofactor is a non-protein chemical compound that is bound to a protein and is required for the protein's biological activity. These proteins are commonly enzymes, and cofactors can be considered "helper molecules" that assist in biochemical transformations....
and a 5'-deoxyadenosyl radical formed in a S-Adenosyl methionine
S-Adenosyl methionine
S-Adenosyl methionine is a common cosubstrate involved in methyl group transfers. SAM was first discovered in Italy by G. L. Cantoni in 1952. It is made from adenosine triphosphate and methionine by methionine adenosyltransferase . Transmethylation, transsulfuration, and aminopropylation are the...
(SAM) activated radical reaction pathway.[1] The generalized reaction is shown below:
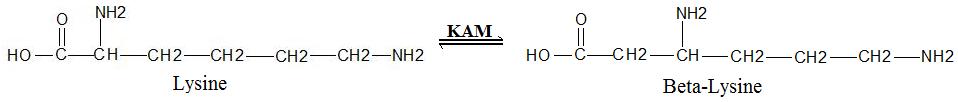
Structure
Shown on the right is the three dimensional structure of the Lysine 2,3-aminomutase protein. The structure was determined by X-ray crystallographyX-ray crystallography
X-ray crystallography is a method of determining the arrangement of atoms within a crystal, in which a beam of X-rays strikes a crystal and causes the beam of light to spread into many specific directions. From the angles and intensities of these diffracted beams, a crystallographer can produce a...
to 2.1 Angstrom resolution and was seen to crystallize as a homotetramer.[2] KAM was first purified and characterized in Clostridium
Clostridium
Clostridium is a genus of Gram-positive bacteria, belonging to the Firmicutes. They are obligate anaerobes capable of producing endospores. Individual cells are rod-shaped, which gives them their name, from the Greek kloster or spindle...
subterminale for studies of Lysine metabolism.
Cofactors

- Pyridoxal phosphate (PLP): Responsible for binding of the amino acid during reaction. The pi-system of this molecule facilitates radical delocalization during formation of an aziridinyl radical. The structure is given below:
- Zinc metalZincZinc , or spelter , is a metallic chemical element; it has the symbol Zn and atomic number 30. It is the first element in group 12 of the periodic table. Zinc is, in some respects, chemically similar to magnesium, because its ion is of similar size and its only common oxidation state is +2...
: Required for coordination between the dimers in the protein. - Iron-sulfur clusterIron-sulfur clusterFor biological Fe-S clusters, see iron-sulfur proteins.Iron-sulfur clusters are ensembles of iron and sulfide centres. Fe-S clusters are most often discussed in the context of the biological role for iron-sulfur proteins. Many Fe-S clusters are known in the area of organometallic chemistry and as...
: A 4 iron-4 sulfur cluster is required for formation of a 5'-deoxyadenosyl radical. This radical then acts as the "stable" radical carrier in the reaction mechanism which transfers the radical to the amino acid.
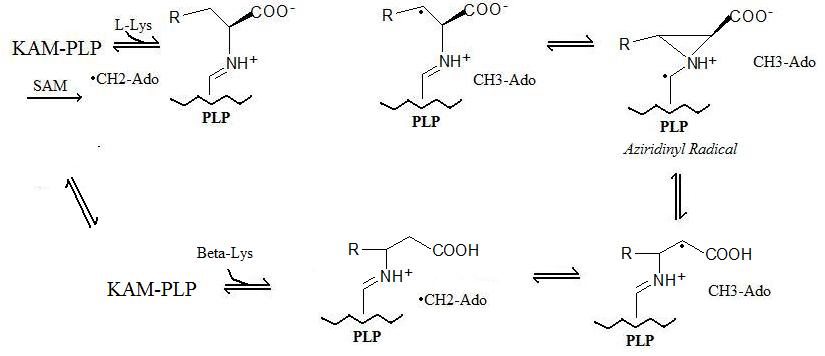
Reaction Mechanism
The generalized reaction takes place in 5 steps:- Radical Formation: A "stable" radical is formed through a radical SAM mechanism in which a S-adenosyl methionine forms a 5'-deoxyadenosyl radical.
- Enzyme Binding: Lysine 2,3-aminomutase binds to pyridoxal phosphate (PLP).
- Amino Acid Binding: The amino acid (Lysine or Beta-Lysine depending on forward or reverse reactions) binds to pyridoxal phosphate.
- Radical Transfer: The 5'-deoxyadenosyl radical is transferred to the amino acid and an aziridinyl radical is formed. In this configuration, the radical is stablizied by the pi-system of pyridoxal phosphate.
- Amino Acid Conversion: In the final step, the new amino acid is formed and the radical is returned to its more stable state on the 5'-deoxyadenosyl.
The reaction mechanism described above is shown below: