
Eukaryotic translation
Encyclopedia
Eukaryotic translation is the process by which messenger RNA
is translated into protein
s in eukaryotes. It consists of initiation, elongation and termination nobelprize.org Translation
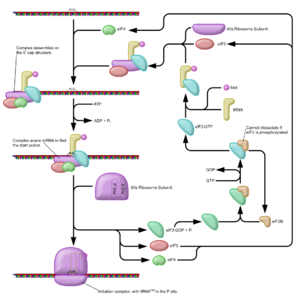
. The protein factors bind the small ribosomal
subunit (also referred to as the 40S
subunit), and these initiation factor
s hold the mRNA in place. The eukaryotic Initiation Factor 3 (eIF3) is associated with the small ribosomal subunit, and plays a role in keeping the large ribosomal subunit from prematurely binding. eIF3 also interacts with the eIF4F complex which consists of three other initiation factors: eIF4A, eIF4E
and eIF4G. eIF4G is a scaffolding protein which directly associates with both eIF3 and the other two components. eIF4E is the cap-binding protein. It is the rate-limiting step of cap-dependent initiation, and is often cleaved from the complex by some viral proteases to limit the cell's ability to translate its own transcripts. This is a method of hijacking the host machinery in favor of the viral (cap-independent) messages. eIF4A is an ATP-dependent RNA helicase, which aids the ribosome in resolving certain secondary structures formed by the mRNA transcript. There is another protein associated with the eIF4F complex called the Poly(A)-binding protein
(PABP), which binds the poly-A tail of most eukaryotic mRNA molecules. This protein has been implicated in playing a role in circularization of the mRNA during translation.
This pre-initiation complex (43S subunit, or the 40S and tRNA) accompanied by the protein factors move along the mRNA chain towards its 3'-end, scanning for the 'start' codon (typically AUG) on the mRNA, which indicates where the mRNA will begin coding for the protein. In eukaryote
s and archaea
, the amino acid
encoded by the start codon is methionine
. The initiator tRNA charged with Met forms part of the ribosomal complex and thus all proteins start with this amino acid (unless it is cleaved away by a protease in subsequent modifications). The Met-charged initiator tRNA is brought to the P-site of the small ribosomal subunit by eukaryotic Initiation Factor 2 (eIF2). It hydrolyzes GTP, and signals for the dissociation of several factors from the small ribosomal subunit which results in the association of the large subunit (or the 60S subunit). The complete ribosome (80S) then commences translation elongation, during which the sequence between the 'start' and 'stop' codons is translated from mRNA into an amino acid sequence—thus a protein is synthesized.
Regulation of protein synthesis is dependent on phosphorylation of initiation factor eIF2 which is a part of the met-tRNAi complex. When large numbers of eIF2 are phosphorylated, protein synthesis is inhibited. This would occur if there is amino acid starvation or there has been a virus infection. However, naturally a small percentage is of this initiation factor is phosphorylated. Another regulator is 4EBP which binds to the initiation factor eIF4E found on the 5’ cap on mRNA stopping protein synthesis. To oppose the effects of the 4EBP growth factors phosphorylate 4EBP reducing its affinity for eIF4E and permitting protein synthesis.
 Elongation is dependent on eukaryotic elongation factors
Elongation is dependent on eukaryotic elongation factors
.
At the end of the initiation step, the mRNA is positioned so that the next codon can be translated during the elongation stage of protein synthesis. The initiator tRNA occupies the P site in the ribosome, and the A site is ready to receive an aminoacyl-tRNA. During chain elongation, each additional amino acid is added to the nascent polypeptide chain in a three-step microcycle. The steps in this microcycle are (1) positioning the correct aminoacyl-tRNA in the A site of the ribosome, (2) forming the peptide bond and (3) shifting the mRNA by one codon relative to the ribosome.
The translation machinery works relatively slowly compared to the enzyme systems that catalyze DNA replication. Proteins in prokaryotes are synthesized at a rate of only 18 amino acid residues per second, whereas bacterial replisomes synthesize DNA at a rate of 1000 nucleotides per second. This difference in rate reflects, in part, the difference between polymerizing four types of nucleotides to make nucleic acids and polymerizing 20 types of amino acids to make proteins. Testing and rejecting incorrect aminoacyl-tRNA molecules takes time and slows protein synthesis.
The rate of transcription in prokaryotes is approximately 55 nucleotides per second, which corresponds to about 18 codons per second, or the same rate at which the mRNA is translated. In bacteria, translation initiation occurs as soon as the 5' end of an mRNA is synthesized, and translation and transcription are coupled. This tight coupling is not possible in eukaryotes because transcription and translation are carried out in separate compartments of the cell (the nucleus and cytoplasm). Eukaryotic mRNA precursors must be processed in the nucleus (e.g. capping, polyadenylation, splicing) before they are exported to the cytoplasm for translation.
. The process is similar to that of prokaryotic termination.
Messenger RNA
Messenger RNA is a molecule of RNA encoding a chemical "blueprint" for a protein product. mRNA is transcribed from a DNA template, and carries coding information to the sites of protein synthesis: the ribosomes. Here, the nucleic acid polymer is translated into a polymer of amino acids: a protein...
is translated into protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
s in eukaryotes. It consists of initiation, elongation and termination nobelprize.org Translation
Initiation
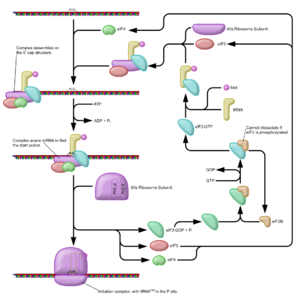
Cap-dependent initiation
Initiation of translation usually involves the interaction of certain key proteins with a special tag bound to the 5'-end of an mRNA molecule, the 5' cap5' cap
The 5' cap is a specially altered nucleotide on the 5' end of precursor messenger RNA and some other primary RNA transcripts as found in eukaryotes. The process of 5' capping is vital to creating mature messenger RNA, which is then able to undergo translation...
. The protein factors bind the small ribosomal
Ribosome
A ribosome is a component of cells that assembles the twenty specific amino acid molecules to form the particular protein molecule determined by the nucleotide sequence of an RNA molecule....
subunit (also referred to as the 40S
40S
40S is the small subunit of eukaryotic ribosomes.It interacts with the internal ribosome entry site of the hepatitis C virus.The following is a list of proteins contained in the 40S ribosome:...
subunit), and these initiation factor
Initiation factor
Initiation factors are proteins that bind to the small subunit of the ribosome during the initiation of translation, a part of protein biosynthesis.They are divided into three major groups:*Prokaryotic initiation factors*Archaeal initiation factors...
s hold the mRNA in place. The eukaryotic Initiation Factor 3 (eIF3) is associated with the small ribosomal subunit, and plays a role in keeping the large ribosomal subunit from prematurely binding. eIF3 also interacts with the eIF4F complex which consists of three other initiation factors: eIF4A, eIF4E
EIF4E
Eukaryotic translation initiation factor 4E, also known as eIF4E, is a protein which in humans is encoded by the eIF4E gene.- Function :...
and eIF4G. eIF4G is a scaffolding protein which directly associates with both eIF3 and the other two components. eIF4E is the cap-binding protein. It is the rate-limiting step of cap-dependent initiation, and is often cleaved from the complex by some viral proteases to limit the cell's ability to translate its own transcripts. This is a method of hijacking the host machinery in favor of the viral (cap-independent) messages. eIF4A is an ATP-dependent RNA helicase, which aids the ribosome in resolving certain secondary structures formed by the mRNA transcript. There is another protein associated with the eIF4F complex called the Poly(A)-binding protein
Poly(A)-binding protein
Poly-binding protein is a RNA-binding protein which binds to the poly tail of mRNA. The poly tail is located on the 3' end of mRNA...
(PABP), which binds the poly-A tail of most eukaryotic mRNA molecules. This protein has been implicated in playing a role in circularization of the mRNA during translation.
This pre-initiation complex (43S subunit, or the 40S and tRNA) accompanied by the protein factors move along the mRNA chain towards its 3'-end, scanning for the 'start' codon (typically AUG) on the mRNA, which indicates where the mRNA will begin coding for the protein. In eukaryote
Eukaryote
A eukaryote is an organism whose cells contain complex structures enclosed within membranes. Eukaryotes may more formally be referred to as the taxon Eukarya or Eukaryota. The defining membrane-bound structure that sets eukaryotic cells apart from prokaryotic cells is the nucleus, or nuclear...
s and archaea
Archaea
The Archaea are a group of single-celled microorganisms. A single individual or species from this domain is called an archaeon...
, the amino acid
Amino acid
Amino acids are molecules containing an amine group, a carboxylic acid group and a side-chain that varies between different amino acids. The key elements of an amino acid are carbon, hydrogen, oxygen, and nitrogen...
encoded by the start codon is methionine
Methionine
Methionine is an α-amino acid with the chemical formula HO2CCHCH2CH2SCH3. This essential amino acid is classified as nonpolar. This amino-acid is coded by the codon AUG, also known as the initiation codon, since it indicates mRNA's coding region where translation into protein...
. The initiator tRNA charged with Met forms part of the ribosomal complex and thus all proteins start with this amino acid (unless it is cleaved away by a protease in subsequent modifications). The Met-charged initiator tRNA is brought to the P-site of the small ribosomal subunit by eukaryotic Initiation Factor 2 (eIF2). It hydrolyzes GTP, and signals for the dissociation of several factors from the small ribosomal subunit which results in the association of the large subunit (or the 60S subunit). The complete ribosome (80S) then commences translation elongation, during which the sequence between the 'start' and 'stop' codons is translated from mRNA into an amino acid sequence—thus a protein is synthesized.
Regulation of protein synthesis is dependent on phosphorylation of initiation factor eIF2 which is a part of the met-tRNAi complex. When large numbers of eIF2 are phosphorylated, protein synthesis is inhibited. This would occur if there is amino acid starvation or there has been a virus infection. However, naturally a small percentage is of this initiation factor is phosphorylated. Another regulator is 4EBP which binds to the initiation factor eIF4E found on the 5’ cap on mRNA stopping protein synthesis. To oppose the effects of the 4EBP growth factors phosphorylate 4EBP reducing its affinity for eIF4E and permitting protein synthesis.
The cap-independent initiation
The best studied example of the cap-independent mode of translation initiation in eukaryotes is the Internal Ribosome Entry Site (IRES) approach. What differentiates cap-independent translation from cap-dependent translation is that cap-independent translation does not require the ribosome to start scanning from the 5' end of the mRNA cap until the start codon. The ribosome can be trafficked to the start site by ITAFs (IRES trans-acting factors) bypassing the need to scan from the 5' end of the untranslated region of the mRNA. This method of translation has been recently discovered, and has found to be important in conditions that require the translation of specific mRNAs, despite cellular stress or the inability to translate most mRNAs. Examples include factors responding to apoptosis, stress-induced responses.Elongation

Eukaryotic elongation factors
Eukaryotic elongation factors are very similar to those in prokaryotes.Elongation in eukaryotes is carried out with two elongation factors: eEF-1 and eEF-2....
.
At the end of the initiation step, the mRNA is positioned so that the next codon can be translated during the elongation stage of protein synthesis. The initiator tRNA occupies the P site in the ribosome, and the A site is ready to receive an aminoacyl-tRNA. During chain elongation, each additional amino acid is added to the nascent polypeptide chain in a three-step microcycle. The steps in this microcycle are (1) positioning the correct aminoacyl-tRNA in the A site of the ribosome, (2) forming the peptide bond and (3) shifting the mRNA by one codon relative to the ribosome.
The translation machinery works relatively slowly compared to the enzyme systems that catalyze DNA replication. Proteins in prokaryotes are synthesized at a rate of only 18 amino acid residues per second, whereas bacterial replisomes synthesize DNA at a rate of 1000 nucleotides per second. This difference in rate reflects, in part, the difference between polymerizing four types of nucleotides to make nucleic acids and polymerizing 20 types of amino acids to make proteins. Testing and rejecting incorrect aminoacyl-tRNA molecules takes time and slows protein synthesis.
The rate of transcription in prokaryotes is approximately 55 nucleotides per second, which corresponds to about 18 codons per second, or the same rate at which the mRNA is translated. In bacteria, translation initiation occurs as soon as the 5' end of an mRNA is synthesized, and translation and transcription are coupled. This tight coupling is not possible in eukaryotes because transcription and translation are carried out in separate compartments of the cell (the nucleus and cytoplasm). Eukaryotic mRNA precursors must be processed in the nucleus (e.g. capping, polyadenylation, splicing) before they are exported to the cytoplasm for translation.
Termination
Termination of elongation is dependent on eukaryotic release factorsEukaryotic release factors
Eukaryotic translation termination factor 1 , also known asTB3-1, is a protein that in humans is encoded by the ETF1 gene.In eukaryotes, there is only one release factor, eRF, which recognizes all three stop codons in place of RF1, RF2, or RF3...
. The process is similar to that of prokaryotic termination.
See also
- Eukaryotic initiation factorEukaryotic initiation factorEukaryotic initiation factors are proteins involved in the initiation phase of eukaryotic translation. They function in forming a complex with the 40S ribosomal subunit and Met-tRNAi called the 43S preinitation complex , recognizing the 5' cap structure of mRNA and recruiting the 43S PIC to mRNA,...
- Eukaryotic elongation factorsEukaryotic elongation factorsEukaryotic elongation factors are very similar to those in prokaryotes.Elongation in eukaryotes is carried out with two elongation factors: eEF-1 and eEF-2....
- Eukaryotic release factorsEukaryotic release factorsEukaryotic translation termination factor 1 , also known asTB3-1, is a protein that in humans is encoded by the ETF1 gene.In eukaryotes, there is only one release factor, eRF, which recognizes all three stop codons in place of RF1, RF2, or RF3...
- Rotavirus translationRotavirus translationRotavirus translation, the process of translating mRNA into proteins, occurs in a different way in Rotaviruses. Unlike the vast majority of cellular proteins in other organisms, in Rotaviruses the proteins are translated from capped but nonpolyadenylated mRNAs...

