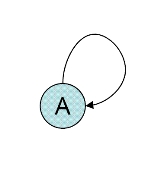
Network motif
Encyclopedia
Network motifs are connectivity-patterns (sub-graphs) that occur much more often than they do in random networks. Most networks studied in biology, ecology and other fields have been found to show a small set of network motifs; surprisingly, in most cases the networks seem to be largely composed of these network motifs, occurring again and again. Each type of network seems to display its own set of characteristic motifs (ecological networks have different motifs than gene regulation networks, etc.). These small circuits can be considered as simple building blocks from which the network is composed. This idea was first presented by Uri Alon
and his group when network motifs were discovered in the gene regulation (transcription) network of the bacteria E. coli
and then in a large set of natural networks. In following work, the network motifs found in E. coli
were discovered in the transcription networks of other bacteria as well as yeast and higher organisms. A distinct set of network motifs were identified in other types of biological networks such as neuronal networks and protein interaction networks.
and yeast, for example, is made of three main motif families, that make up almost the entire network. The leading hypothesis is that the network motif were independently selected by evolutionary processes in a converging manner, since the creation or elimination of regulatory interactions is fast on evolutionary time scale, relative to the rate at which genes change, Furthermore, experiments on the dynamics generated by network motifs in living cells indicate that they have characteristic dynamical functions. This suggests that the network motif serve as building blocks in gene regulatory networks that are beneficial to the organism.
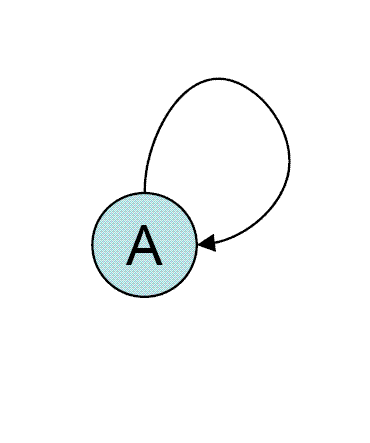 One of simplest and most abundant network motifs in E. coli
One of simplest and most abundant network motifs in E. coli
is negative auto-regulation in which a transcription factor (TF) represses its own transcription. This motif was shown to perform two important functions. The first function is response acceleration. NAR was shown to speed-up the response to signals both theoretically and experimentally. This was first shown in a synthetic transcription network and later on in the natural context in the SOS DNA repair system of E .coli. The second function is increased stability of the auto-regulated gene product concentration against stochastic noise, thus reducing variations in protein levels between different cells.
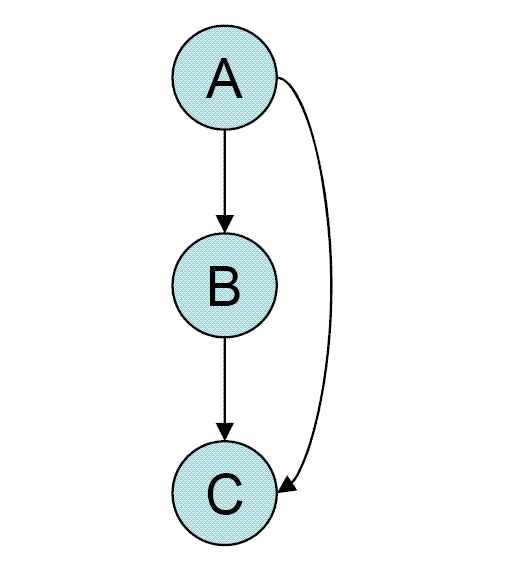 This motif is commonly found in many gene systems and organisms. The motif consists of three genes and three regulatory interactions. The target gene Z is regulated by 2 TFs X and Y and in addition TF Y is also regulated TF X . Since each of the regulatory interactions may either be positive or negative there are possibly eight types of FFL motifs. Two of those eight types: the coherent type 1 FFL (C1-FFL) (where all interactions are positive) and the incoherent type 1 FFL (I1-FFL) (X activates Z and also activates Y which represses Z) are found much more frequently in the transcription network of E. coli
This motif is commonly found in many gene systems and organisms. The motif consists of three genes and three regulatory interactions. The target gene Z is regulated by 2 TFs X and Y and in addition TF Y is also regulated TF X . Since each of the regulatory interactions may either be positive or negative there are possibly eight types of FFL motifs. Two of those eight types: the coherent type 1 FFL (C1-FFL) (where all interactions are positive) and the incoherent type 1 FFL (I1-FFL) (X activates Z and also activates Y which represses Z) are found much more frequently in the transcription network of E. coli
and yeast than the other six types. In addition to the structure of the circuitry the way in which the signals from X and Y are integrated by the Z promoter should also be considered. In most of the cases the FFL is either an AND gate (X and Y are required for Z activation) or OR gate (either X or Y are sufficient for Z activation) but other input function are also possible.
. This means that this motif can provide pulse filtration in which short pulses of signal will not generate a response but persistent signals will generate a response after short delay. The shut off of the output when a persistent pulse is ended will be fast. The opposite behavior emerges in the case of a sum gate with fast response and delayed shut off as was demonstrated in the flagella system of E. coli
.
.
In the literature, Multiple-input modules (MIM) arose as a generalization of SIM. However, the precise definitions of SIM and MIM have been a source of inconsistency. There are attempts to provide orthogonal definitions for canonical motifs in biological networks and algorithms to enumerate them, especially SIM, MIM and Bi-Fan (2x2 MIM).
in various systems such as carbon utilization, anaerobic growth, stress response and others. In order to better understand the function of this motif one has to obtain more information about the way the multiple inputs are integrated by the genes. Kaplan et al. has mapped the input functions of the sugar utilization genes in E. coli
, showing diverse shapes.
Most analyses of motif function are carried out looking at the motif operating in isolation. Recent research provides good evidence that network context, i.e. the connections of the motif to the rest of the network, are too important to draw inferences on function from local structure only — the cited paper also reviews the criticisms and alternative explanations for the observed data. Yet another recent work suggests that certain topological features of biological networks naturally give rise to the common appearance of canonical motifs, thereby questioning whether frequencies of occurrences are reasonable evidence that the structures of motifs are selected for their functional contribution to the operation of networks.
Uri Alon
Uri Alon is a Professor and Systems Biologist at the Weizmann Institute of Science. His highly cited research investigates gene expression, network motifs and the design principles of biological networks in Escherichia coli and other organisms using both computational biology and traditional...
and his group when network motifs were discovered in the gene regulation (transcription) network of the bacteria E. coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
and then in a large set of natural networks. In following work, the network motifs found in E. coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
were discovered in the transcription networks of other bacteria as well as yeast and higher organisms. A distinct set of network motifs were identified in other types of biological networks such as neuronal networks and protein interaction networks.
Network motifs in gene regulation networks
Much experimental work has been devoted to understanding network motifs in gene regulatory networks. These networks control which genes are expressed in the cell in response to biological signals. The network is defined such that genes are nodes, and directed edges represent the control of one gene by a transcription factor (regulatory protein that binds DNA) encoded by another gene. Thus, network motifs are patterns of genes regulating each others transcription rate. When analyzing transcription networks, it is seen that the same network motifs appear again and again in diverse organisms from bacteria to human. The transcription network of E. coliEscherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
and yeast, for example, is made of three main motif families, that make up almost the entire network. The leading hypothesis is that the network motif were independently selected by evolutionary processes in a converging manner, since the creation or elimination of regulatory interactions is fast on evolutionary time scale, relative to the rate at which genes change, Furthermore, experiments on the dynamics generated by network motifs in living cells indicate that they have characteristic dynamical functions. This suggests that the network motif serve as building blocks in gene regulatory networks that are beneficial to the organism.
The function of network motifs
The functions associated with common network motifs in transcription networks were explored and demonstrated by several research projects both theoretically and experimentally. Below are some of the most common network motifs and their associated function.Negative auto-regulation (NAR)

Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
is negative auto-regulation in which a transcription factor (TF) represses its own transcription. This motif was shown to perform two important functions. The first function is response acceleration. NAR was shown to speed-up the response to signals both theoretically and experimentally. This was first shown in a synthetic transcription network and later on in the natural context in the SOS DNA repair system of E .coli. The second function is increased stability of the auto-regulated gene product concentration against stochastic noise, thus reducing variations in protein levels between different cells.
Positive auto-regulation (PAR)
Positive auto-regulation (PAR) occurs when a transcription factor enhances its own rate of production. Opposite to the NAR motif this motif slows the response time compared to simple regulation. In the case of a strong PAR the motif may lead to a bimodal distribution of protein levels in cell populations.Feed-forward loops (FFL)

Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
and yeast than the other six types. In addition to the structure of the circuitry the way in which the signals from X and Y are integrated by the Z promoter should also be considered. In most of the cases the FFL is either an AND gate (X and Y are required for Z activation) or OR gate (either X or Y are sufficient for Z activation) but other input function are also possible.
Coherent type 1 FFL (C1-FFL)
The C1-FFL with an AND gate was shown to have a function of a ‘sign-sensitive delay’ element and a persistence detector both theoretically and experimentally with the arabinose system of E. coliEscherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
. This means that this motif can provide pulse filtration in which short pulses of signal will not generate a response but persistent signals will generate a response after short delay. The shut off of the output when a persistent pulse is ended will be fast. The opposite behavior emerges in the case of a sum gate with fast response and delayed shut off as was demonstrated in the flagella system of E. coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
.
Incoherent type 1 FFL (I1-FFL)
The I1-FFL is a pulse generator and response accelerator. The two signal pathways of the I1-FFL acts in opposite directions where one pathway activates Z and the other represses it. When the repression is complete this leads to a pulse-like dynamics. It was also demonstrated experimentally that the I1-FFL can serve as response accelerator in a way which is similar to the NAR motif. The difference is that the I1-FFL can speed-up the response of any gene and not necessarily a transcription factor gene. Recently additional function was assigned to the I1-FFL network motif: it was shown both theoretically and experimentally that the I1-FFL can generate non-monotonic input function in both a synthetic and native systems.Multi-output FFLs
In some cases the same regulators X and Y regulate several Z genes of the same system. By adjusting the strength of the interactions this motif was shown to determine the temporal order of gene activation. This was demonstrated experimentally in the flagella system of E. coliEscherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
.
Single-input modules (SIM)
This motif occurs when a single regulator regulates a set of genes with no additional regulation. This is useful when the genes are cooperatively carrying out a specific function and therefore always need to be activated in a synchronized manner. By adjusting the strength of the interactions it can create temporal expression program of the genes it regulates.In the literature, Multiple-input modules (MIM) arose as a generalization of SIM. However, the precise definitions of SIM and MIM have been a source of inconsistency. There are attempts to provide orthogonal definitions for canonical motifs in biological networks and algorithms to enumerate them, especially SIM, MIM and Bi-Fan (2x2 MIM).
Dense overlapping regulons (DOR)
This motif occurs in the case that several regulators combinatorially control a set of genes with diverse regulatory combinations. This motif was found in E. coliEscherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
in various systems such as carbon utilization, anaerobic growth, stress response and others. In order to better understand the function of this motif one has to obtain more information about the way the multiple inputs are integrated by the genes. Kaplan et al. has mapped the input functions of the sugar utilization genes in E. coli
Escherichia coli
Escherichia coli is a Gram-negative, rod-shaped bacterium that is commonly found in the lower intestine of warm-blooded organisms . Most E. coli strains are harmless, but some serotypes can cause serious food poisoning in humans, and are occasionally responsible for product recalls...
, showing diverse shapes.
Activity motifs
An interesting generalization of the network-motifs, activity motifs are over occurring patterns that can be found when nodes and edges in the network are annotated with quantitative features. For instance, when edges in a metabolic pathways are annotated with the magnitude or timing of the corresponding gene expression, some patterns are over occurring given the underlying network structure.Criticism
An assumption (sometimes more sometimes less implicit) behind the preservation of a topological sub-structure is that it is of a particular functional importance. This assumption has recently been questioned. Some authors have argued that motifs, like bi-fan motifs, might show a variety depending on the network context, and therefore, structure of the motif does not necessarily determine function. Network structure certainly does not always indicate function; this is an idea that has been around for some time, for an example see the Sin operon.Most analyses of motif function are carried out looking at the motif operating in isolation. Recent research provides good evidence that network context, i.e. the connections of the motif to the rest of the network, are too important to draw inferences on function from local structure only — the cited paper also reviews the criticisms and alternative explanations for the observed data. Yet another recent work suggests that certain topological features of biological networks naturally give rise to the common appearance of canonical motifs, thereby questioning whether frequencies of occurrences are reasonable evidence that the structures of motifs are selected for their functional contribution to the operation of networks.

