
DEAD box
Encyclopedia
DEAD box proteins are involved in an assortment of metabolic processes that involve RNA. They are highly conserved in nine domains and can be found in both prokaryotes and eukaryotes, but not all. Many organisms, including humans, contain DEAD-box helicase
s, which are involved in RNA
metabolism
.
and contains the amino acid sequence D-E-A-D (asp-glu-ala-asp), which gave this family of proteins the name “DEAD box”. Motif 1, motif II, the Q motif, and motif VI are all needed for ATP binding and hydrolysis, while motifs, 1a, 1b, III, IV, and V may be involved in intramolecular rearrangements and RNA interaction.

. They are all quite distinct from one another and there is not one protein that belongs to more than one of these families. It is thought that each family has a specific role in RNA metabolism, for example both DEAD box and DEAH box proteins NTPase activities become stimulated by RNA, but DEAD box proteins use ATP and DEAH does not.
, release of the mRNA, and recycling of the spliceosome complex9.
Helicase
Helicases are a class of enzymes vital to all living organisms. They are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands using energy derived from ATP hydrolysis.-Function:Many cellular processes Helicases are a...
s, which are involved in RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
metabolism
Metabolism
Metabolism is the set of chemical reactions that happen in the cells of living organisms to sustain life. These processes allow organisms to grow and reproduce, maintain their structures, and respond to their environments. Metabolism is usually divided into two categories...
.
DEAD Box Family
DEAD box proteins were first brought to attention in the late 1980s in a study that looked at a group of NTPases that were similar in sequence to the eIF4A RNA helicase sequence. The results of this study showed that these proteins (p68,SrmB, MSS116, vasa, PL10, mammalian eIF4A, yeast eIF4A) involved in RNA metabolism had several common elements. There were nine common sequences found to be conserved amongst the studied species, which is an important criterion of the DEAD box family. The nine conserved motifs are as follows, Q-motif, motif 1, motif 1a, motif 1b, motif II, motif III, motif IV, motif V, and motif VI, as shown in the figure. Motif II is also known as the Walker B motifWalker motifs
The Walker A and Walker B motifs are protein sequence motifs. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.-Walker A motif:...
and contains the amino acid sequence D-E-A-D (asp-glu-ala-asp), which gave this family of proteins the name “DEAD box”. Motif 1, motif II, the Q motif, and motif VI are all needed for ATP binding and hydrolysis, while motifs, 1a, 1b, III, IV, and V may be involved in intramolecular rearrangements and RNA interaction.

Related Families
The DEAH and Ski families have had proteins that have been identified to be related to the DEAD box family. These two relatives have a few particularly unique motifs that are conserved within their own family. DEAD box, DEAH, and the Ski families are all referred to as DExD/H proteinsDExD/H box proteins
DEAD box, DEAH, and the Ski families of proteins are all referred to as DExD/H box proteins. They are all quite distinct from one another and there is not one protein that belongs to more than one of these families...
. They are all quite distinct from one another and there is not one protein that belongs to more than one of these families. It is thought that each family has a specific role in RNA metabolism, for example both DEAD box and DEAH box proteins NTPase activities become stimulated by RNA, but DEAD box proteins use ATP and DEAH does not.
Biological Functions
DEAD box proteins are considered to be RNA helicases and many have been found to be required in cellular processes such as pre-mRNA processing and rearranging of ribonucleoproteins (RNP) complexes.Pre-mRNA Splicing
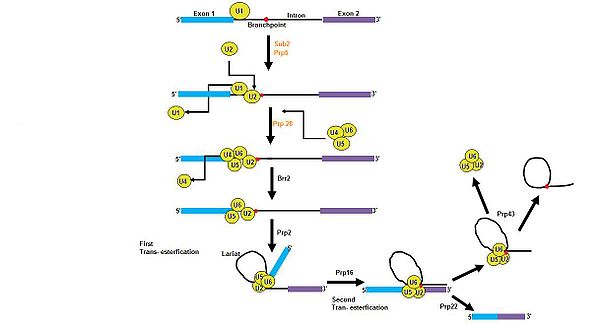
Pre-mRNA splicing requires rearrangements of five large RNP complexes, which are snRNPs U1, U2, U4, U5, and U6. DEAD box proteins are helicases that perform unwinding in an energy dependent approach and are able to perform these snRNP rearrangements in a quick and efficient manner. There are three DEAD box proteins in the yeast system, Sub2, Prp28, and Prp5, and have been proven to be required for in vivo splicing. Prp5 has been shown to assist in a conformational rearrangement of U2 snRNA, which makes the branch point recognition sequence of U2 available to bind the branch point sequence. Prp28 may have a role in recognizing the 5’ splice site and does not display RNA helicase activity, suggesting that other factors must be present in order to activate Prp28. DExD/H proteins have also been found to be required components in pre- mRNA splicing, in particular the DEAH proteins, Prp2, Prp16, Prp22, Prp43, and Brr213. As shown in the figure, DEAD box proteins are needed in the initial steps of spliceosome formation, while DEAH box proteins are indirectly required for the transesterificationsTransesterification
In organic chemistry, transesterification is the process of exchanging the organic group R″ of an ester with the organic group R′ of an alcohol. These reactions are often catalyzed by the addition of an acid or base catalyst...
, release of the mRNA, and recycling of the spliceosome complex9.

Translation Initiation
The eIF4A translation initiation factor was the first DEAD box protein found to have a RNA dependent ATPase activity. It has been proposed that this abundant protein helps in unwinding the secondary structure in the 5’-untranslated region. This can inhibit the scanning process of the small ribosomal subunit, if not unwound. Ded1 is another DEAD box protein that is also needed for translation initiation, but its exact role in this process is still obscure. Vasa, a DEAD box protein highly related to Ded1 plays a part in translation initiation by interacting with initiation factor 2 (IF2).See also
- DEAD/DEAH box helicaseDEAD/DEAH box helicaseThe DEAD/DEAH box helicases are a family of proteins whose purpose is to unwind nucleic acids. The DEAD box helicases are involved in various aspects of RNA metabolism, including nuclear transcription, pre mRNA splicing, ribosome biogenesis, nucleocytoplasmic transport, translation, RNA decay and...
- Ski 2 helicase
- Walker A motif
- RNA helicase
- DDX3XDDX3XATP-dependent RNA helicase DDX3X is an enzyme that in humans is encoded by the DDX3X gene.-Further reading:...

