Cot analysis
Encyclopedia
C0t analysis, also known as DNA reassociation kinetics, is a biochemical technique that measures how much repetitive DNA is in a genome
. It is used to study genome
structure and organization and has also been used to simplify the sequencing of genomes that contain large amounts of repetitive sequence.
into the single stranded-form, and then slowly cooling it, so the strands can pair back together. While the sample is cooling, measurements are taken of how much of the DNA is base pair
ed at each temperature.
The amount of single and double-stranded DNA is measured by rapidly diluting the sample, which slows reassociation, and then binding the DNA to a hydroxylapatite
column. The column is first washed with a low concentration of sodium phosphate buffer, which elutes the single-stranded DNA, and then with high concentrations of phosphate, which elutes the double stranded DNA. The amount of DNA in these two solutions is then measured using a spectrophotometer.
strand to reform a double helix, common sequences renature more rapidly than rare sequences. Indeed, the rate at which a sequence will reassociate is proportional
to the number of copies of that sequence in the DNA sample. A sample with a highly-repetitive sequence will renature rapidly, while complex sequences will renature slowly.
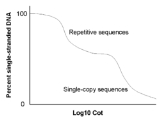
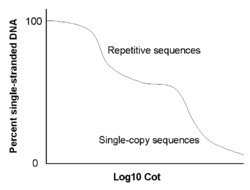
However, instead of simply measuring the percentage of double-stranded DNA versus time, the amount of renaturation is measured relative to a C0t value. The C0t value is the product of C0 (the initial concentration of DNA), t (time in seconds), and a constant that depends on the concentration of cations in the buffer. Repetitive DNA will renature at low C0t values, while complex and unique DNA sequences will renature at high C0t values.
sequence
s that dominate many eukaryotic genomes from "gene-rich" single/low-copy sequences. This allows DNA sequencing to concentrate on the parts of the genome that are most informative and interesting, which will speed up the discovery of new genes and make the process more efficient.
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
. It is used to study genome
Genome
In modern molecular biology and genetics, the genome is the entirety of an organism's hereditary information. It is encoded either in DNA or, for many types of virus, in RNA. The genome includes both the genes and the non-coding sequences of the DNA/RNA....
structure and organization and has also been used to simplify the sequencing of genomes that contain large amounts of repetitive sequence.
Procedure
The procedure involves heating a sample of genomic DNA until it denaturesDenaturation (biochemistry)
Denaturation is a process in which proteins or nucleic acids lose their tertiary structure and secondary structure by application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent , or heat...
into the single stranded-form, and then slowly cooling it, so the strands can pair back together. While the sample is cooling, measurements are taken of how much of the DNA is base pair
Base pair
In molecular biology and genetics, the linking between two nitrogenous bases on opposite complementary DNA or certain types of RNA strands that are connected via hydrogen bonds is called a base pair...
ed at each temperature.
The amount of single and double-stranded DNA is measured by rapidly diluting the sample, which slows reassociation, and then binding the DNA to a hydroxylapatite
Hydroxylapatite
Hydroxylapatite, also called hydroxyapatite , is a naturally occurring mineral form of calcium apatite with the formula Ca53, but is usually written Ca1062 to denote that the crystal unit cell comprises two entities. Hydroxylapatite is the hydroxyl endmember of the complex apatite group...
column. The column is first washed with a low concentration of sodium phosphate buffer, which elutes the single-stranded DNA, and then with high concentrations of phosphate, which elutes the double stranded DNA. The amount of DNA in these two solutions is then measured using a spectrophotometer.

Analysis
Since a sequence of single-stranded DNA needs to find its complementaryComplementarity (molecular biology)
In molecular biology, complementarity is a property of double-stranded nucleic acids such as DNA, as well as DNA:RNA duplexes. Each strand is complementary to the other in that the base pairs between them are non-covalently connected via two or three hydrogen bonds...
strand to reform a double helix, common sequences renature more rapidly than rare sequences. Indeed, the rate at which a sequence will reassociate is proportional
Proportionality (mathematics)
In mathematics, two variable quantities are proportional if one of them is always the product of the other and a constant quantity, called the coefficient of proportionality or proportionality constant. In other words, are proportional if the ratio \tfrac yx is constant. We also say that one...
to the number of copies of that sequence in the DNA sample. A sample with a highly-repetitive sequence will renature rapidly, while complex sequences will renature slowly.
However, instead of simply measuring the percentage of double-stranded DNA versus time, the amount of renaturation is measured relative to a C0t value. The C0t value is the product of C0 (the initial concentration of DNA), t (time in seconds), and a constant that depends on the concentration of cations in the buffer. Repetitive DNA will renature at low C0t values, while complex and unique DNA sequences will renature at high C0t values.
Application to genome sequencing
Cot filtration is a technique that uses the principles of DNA renaturation kinetics to separate the repetitive DNARepeated sequence (DNA)
In the study of DNA sequences, one can distinguish two main types of repeated sequence:*Tandem repeats:**Satellite DNA**Minisatellite**Microsatellite*Interspersed repeats:**SINEs...
sequence
Sequence
In mathematics, a sequence is an ordered list of objects . Like a set, it contains members , and the number of terms is called the length of the sequence. Unlike a set, order matters, and exactly the same elements can appear multiple times at different positions in the sequence...
s that dominate many eukaryotic genomes from "gene-rich" single/low-copy sequences. This allows DNA sequencing to concentrate on the parts of the genome that are most informative and interesting, which will speed up the discovery of new genes and make the process more efficient.
History
It was first developed and utilized by Roy Britten and his colleagues at the Carnegie Institution of Washington in the 1960s. Of particular note, it was through Cot analysis that the redundant (repetitive) nature of eukaryotic genomes was first discovered. However, it wasn't until the breakthrough DNA reassociation kinetics experiments of Britten and his colleagues that it was shown that not all DNA coded for genes. In fact, their experiments demonstrated that the majority of eukaryotic genomic DNA is composed of repetitive, non-coding elements.External links
- Cot Analysis: An Overview Mississippi Genome Exploration Laboratory

