
Biochemical systems theory
Encyclopedia
Biochemical systems theory is a mathematical model
ling framework for biochemical systems, based on ordinary differential equation
s (ODE), in which biochemical processes
are represented using power-law expansions in the variables of the system
.
This framework, which became known as Biochemical Systems Theory, has been developed since the 1960s by Michael Savageau and others for the systems analysis
of biochemical processes. According to Cornish-Bowden (2007) they "regarded this as a general theory
of metabolic control, which includes both metabolic control analysis and flux-oriented theory as special cases".
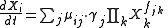
where Xi represents one of the nd variables of the model (metabolite concentrations, protein concentrations or levels of gene expression). j represents the nf biochemical processes affecting the dynamics of the specie. On the other hand,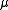 ij (stoichiometric coefficient),
ij (stoichiometric coefficient), 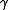 j (rate constants) and fjk (kinetic orders) are two different kinds of parameters defining the dynamics of the system.
j (rate constants) and fjk (kinetic orders) are two different kinds of parameters defining the dynamics of the system.
The principal difference of power-law models
with respect to other ODE models used in biochemical systems is that the kinetic orders can be non-integer numbers. A kinetic order can have even negative value when inhibition is modelled. In this way, power-law models have a higher flexibility to reproduce the non-linearity of biochemical systems.
Models using power-law expansions have been used during the last 35 years to model and analyse several kinds of biochemical systems including metabolic networks, genetic networks and recently in cell signalling.
Scientific articles:
Mathematical model
A mathematical model is a description of a system using mathematical concepts and language. The process of developing a mathematical model is termed mathematical modeling. Mathematical models are used not only in the natural sciences and engineering disciplines A mathematical model is a...
ling framework for biochemical systems, based on ordinary differential equation
Differential equation
A differential equation is a mathematical equation for an unknown function of one or several variables that relates the values of the function itself and its derivatives of various orders...
s (ODE), in which biochemical processes
Biochemistry
Biochemistry, sometimes called biological chemistry, is the study of chemical processes in living organisms, including, but not limited to, living matter. Biochemistry governs all living organisms and living processes...
are represented using power-law expansions in the variables of the system
System
System is a set of interacting or interdependent components forming an integrated whole....
.
This framework, which became known as Biochemical Systems Theory, has been developed since the 1960s by Michael Savageau and others for the systems analysis
Systems analysis
Systems analysis is the study of sets of interacting entities, including computer systems analysis. This field is closely related to requirements analysis or operations research...
of biochemical processes. According to Cornish-Bowden (2007) they "regarded this as a general theory
General Theory
General theory may refer to:*Generalized Theory of Gravitation*General Theory of Relativity*General Systems Theory*Generalized cohomology theory*General theory of collaboration...
of metabolic control, which includes both metabolic control analysis and flux-oriented theory as special cases".
Representation
The dynamics of a species is represented by a differential equation with the structure: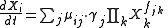
where Xi represents one of the nd variables of the model (metabolite concentrations, protein concentrations or levels of gene expression). j represents the nf biochemical processes affecting the dynamics of the specie. On the other hand,
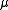 ij (stoichiometric coefficient),
ij (stoichiometric coefficient), 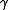 j (rate constants) and fjk (kinetic orders) are two different kinds of parameters defining the dynamics of the system.
j (rate constants) and fjk (kinetic orders) are two different kinds of parameters defining the dynamics of the system.The principal difference of power-law models
Mathematical model
A mathematical model is a description of a system using mathematical concepts and language. The process of developing a mathematical model is termed mathematical modeling. Mathematical models are used not only in the natural sciences and engineering disciplines A mathematical model is a...
with respect to other ODE models used in biochemical systems is that the kinetic orders can be non-integer numbers. A kinetic order can have even negative value when inhibition is modelled. In this way, power-law models have a higher flexibility to reproduce the non-linearity of biochemical systems.
Models using power-law expansions have been used during the last 35 years to model and analyse several kinds of biochemical systems including metabolic networks, genetic networks and recently in cell signalling.
Literature
Books:- M.A. Savageau, Biochemical systems analysis: a study of function and design in molecular biology, Reading, MA, Addison–Wesley, 1976.
- E.O. Voit (ed), Canonical Nonlinear Modeling. S-System Approach to Understanding Complexity, Van Nostrand Reinhold, NY, 1991.
- E.O. Voit, Computational Analysis of Biochemical Systems. A Practical Guide for Biochemists and Molecular Biologists, Cambridge University Press, Cambridge, U.K., 2000.
- N.V. Torres and E.O. Voit, Pathway Analysis and Optimization in Metabolic Engineering, Cambridge University Press, Cambridge, U.K., 2002.
Scientific articles:
- M.A. Savageau, Biochemical systems analysis: I. Some mathematical properties of the rate law for the component enzymatic reactions in: J. Theor. Biol. 25, pp. 365–369, 1969.
- M.A. Savageau, Development of fractal kinetic theory for enzyme-catalysed reactions and implications for the design of biochemical pathways in: Biosystems 47(1-2), pp. 9–36, 1998.
- M.R. Atkinson et al., Design of gene circuits using power-law models, in: Cell 113, pp. 597–607, 2003.
- F. Alvarez-Vasquez et al., Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae, Nature 27, pp. 433(7024), pp. 425–30, 2005.
- J. Vera et al., Power-Law models of signal transduction pathways in: Cellular Signalling doi:10.1016/j.cellsig.2007.01.029), 2007.
- Eberhart O. Voit, Applications of Biochemical Systems Theory, 2006.
External links
- Biochemical Systems Theory an introduction,
- http://web.udl.es/Biomath/PowerLaw/
- A Web on Power-law Models for Biochemical Systems

