
Zinc finger
Encyclopedia
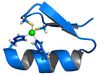
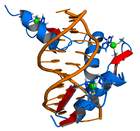
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
structural motif
Structural motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a supersecondary structure, which appears also in a variety of other molecules...
s that can coordinate one or more zinc
Zinc
Zinc , or spelter , is a metallic chemical element; it has the symbol Zn and atomic number 30. It is the first element in group 12 of the periodic table. Zinc is, in some respects, chemically similar to magnesium, because its ion is of similar size and its only common oxidation state is +2...
ion
Ion
An ion is an atom or molecule in which the total number of electrons is not equal to the total number of protons, giving it a net positive or negative electrical charge. The name was given by physicist Michael Faraday for the substances that allow a current to pass between electrodes in a...
s to help stabilize their folds. They can be classified into several different structural families (zinc finger proteins) and typically function as interaction modules that bind DNA
DNA
Deoxyribonucleic acid is a nucleic acid that contains the genetic instructions used in the development and functioning of all known living organisms . The DNA segments that carry this genetic information are called genes, but other DNA sequences have structural purposes, or are involved in...
, RNA
RNA
Ribonucleic acid , or RNA, is one of the three major macromolecules that are essential for all known forms of life....
, proteins, or small molecules. The name "zinc finger" was originally coined to describe the finger-like appearance of a diagram showing the hypothesized structure of the repeated unit in Xenopus laevis transcription factor IIIA
GTF3A
Transcription factor IIIA is a protein that in humans is encoded by the GTF3A gene....
.
Classes
Zinc fingers coordinate zinc ions with a combination of cysteineCysteine
Cysteine is an α-amino acid with the chemical formula HO2CCHCH2SH. It is a non-essential amino acid, which means that it is biosynthesized in humans. Its codons are UGU and UGC. The side chain on cysteine is thiol, which is polar and thus cysteine is usually classified as a hydrophilic amino acid...
and histidine
Histidine
Histidine Histidine, an essential amino acid, has a positively charged imidazole functional group. It is one of the 22 proteinogenic amino acids. Its codons are CAU and CAC. Histidine was first isolated by German physician Albrecht Kossel in 1896. Histidine is an essential amino acid in humans...
residues. They can be classified by the type and order of these zinc coordinating residues (e.g., Cys2His2, Cys4, and Cys6). A more systematic method classifies them into different "fold groups" based on the overall shape of the protein backbone in the folded domain. The most common "fold groups" of zinc fingers are the Cys2His2-like (the "classic zinc finger"), treble clef, and zinc ribbon.
The following table shows the different structures and their key features:
| Fold Group | Representative structure | Ligand placement |
|---|---|---|
| Cys2His2 | Two ligands form a knuckle and two more form the c terminus of a helix. | |
| Gag knuckle | Two ligands form a knuckle and two more form a short helix or loop. | |
| Treble clef | Two ligands form a knuckle and two more form the N terminus of a helix. | |
| Zinc ribbon | Two ligands each form two knuckles. | |
| Zn2/Cys6 | Two ligands form the N terminus of a helix and two more form a loop. | |
| TAZ2 domain like | Two ligands form the termini of two helices. |
Cys2His2
The Cys2His2-like fold group is by far the best-characterized class of zinc fingers and are extremely common in mammalian transcription factors. These domains adopt a simple ββα fold and have the amino acid Sequence motifSequence motif
In genetics, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and has, or is conjectured to have, a biological significance...
:
X2-Cys-X2,4-Cys-X12-His-X3,4,5-His
This class of zinc fingers can have a variety of functions such as binding RNA and mediating protein-protein interactions, but is best known for its role in sequence-specific DNA-binding proteins such as Zif268
Zif268
EGR-1 also known as Zif268 or NGFI-A is a protein that in humans is encoded by the EGR1 gene....
. In such proteins, individual zinc finger domains typically occur as tandem repeats with two, three, or more fingers comprising the DNA-binding domain of the protein. These tandem arrays can bind in the major groove of DNA and are typically spaced at 3-bp intervals. The α-helix of each domain (often called the "recognition helix") can make sequence-specific contacts to DNA bases; residues from a single recognition helix can contact 4 or more bases to yield an overlapping pattern of contacts with adjacent zinc fingers.
Gag-knuckle
This fold group is defined by two short β-strands connected by a turn (zinc knuckle) followed by a short helix or loop and resembles the classical Cys2His2 motif with a large portion of the helix and β-hairpin truncated.The retroviral nucleocapsid (NC) protein from HIV and other related retroviruses are examples of proteins possessing these motifs. The gag-knuckle zinc finger in the HIV NC protein is the target of a class of drugs known as zinc finger inhibitor
Zinc finger inhibitor
Zinc finger inhibition is the process by which the synthesis of zinc fingers is blocked. Zinc finger inhibitors have been tested for their efficacy in treating AIDS and HIV....
s.
Treble-clef
The treble-clef motif consists of a β-hairpin at the N-terminus and an α-helix at the C-terminus that each contribute two ligands for zinc binding, although a loop and a second β-hairpin of varying length and conformation can be present between the N-terminal β-hairpin and the C-terminal α-helix. These fingers are present in a diverse group of proteins that frequently do not share sequence or functional similarity with each other. The best-characterized proteins containing treble-clef zinc fingers are the nuclear hormone receptors.Zinc Ribbon
The zinc ribbon fold is characterised by two beta-hairpins forming two structurally similar zinc-binding sub-sites.Zn2/Cys6
The canonical members of this class contain a binuclear zinc cluster in which two zinc ions are bound by six cysteineCysteine
Cysteine is an α-amino acid with the chemical formula HO2CCHCH2SH. It is a non-essential amino acid, which means that it is biosynthesized in humans. Its codons are UGU and UGC. The side chain on cysteine is thiol, which is polar and thus cysteine is usually classified as a hydrophilic amino acid...
residues. These zinc fingers can be found in several transcription factors including the yeast Gal4 protein.
Engineered zinc finger arrays
Generating arrays of engineered Cys2His2 zinc fingers is the most developed method for creating proteins capable of targeting desired genomic DNA sequences. The majority of engineered zinc finger arrays are based on the zinc finger domain of the murine transcription factor Zif268, although some groups have used zinc finger arrays based on the human transcription factor SP1. Zif268 has three individual zinc finger motifs that collectively bind a 9 bp sequence with high affinity.The structure of this protein bound to DNA was solved in 1991 and stimulated a great deal of research into engineered zinc finger arrays. In 1994 and 1995, a number of groups used phage display
Phage display
Phage display is a method for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. Phage Display was originally invented by George P...
to alter the specificity of a single zinc finger of Zif268.
Carlos F. Barbas et al. also reported the development of zinc finger technology in the patent literature and have been granted a number of patents that have been important for the commercial development of zinc finger technology. Typical engineered zinc finger arrays have between 3 and 6 individual zinc finger motifs and bind target sites ranging from 9 basepairs to 18 basepairs in length. Arrays with 6 zinc finger motifs are particularly attractive because they bind a target site that is long enough to have a good chance of being unique in a mammalian genome.
There are two main methods currently used to generate engineered zinc finger arrays, modular assembly and a bacterial selection system, and there is some debate about which method is best suited for most applications.
Modular assembly
The most straightforward method to generate new zinc finger arrays is to combine smaller zinc finger "modules" of known specificity. The structure of the zinc finger protein Zif268 bound to DNA described by Pavletich and Pabo in their 1991 publication has been key to much of this work and describes the concept of obtaining fingers for each of the 64 possible base pair triplets and then mixing and matching these fingers to design proteins with any desired sequence specificity.The most common modular assembly process involves combining separate zinc fingers that can each recognize a 3 basepair DNA sequence to generate 3-finger, 4-, 5-, or 6-finger arrays that recognize target sites ranging from 9 basepairs to 18 basepairs in length. Another method uses 2-finger modules to generate zinc finger arrays with up to six individual zinc fingers.
The Barbas Laboratory of The Scripps Research Institute used phage display
Phage display
Phage display is a method for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. Phage Display was originally invented by George P...
to develop and characterize zinc finger domains that recognize most DNA triplet sequences
while another group isolated and characterized individual fingers from the human genome.
A potential drawback with modular assembly in general is that specificities of individual zinc finger can overlap and can depend on the context of the surrounding zinc fingers and DNA. A recent study demonstrated that a high proportion of 3-finger zinc finger arrays generated by modular assembly fail to bind their intended target with sufficient affinity in a bacterial two-hybrid assay and fail to function as zinc finger nucleases, but the success rate was somewhat higher when sites of the form GNNGNNGNN were targeted. A subsequent study used modular assembly to generate zinc finger nucleases with both 3-finger arrays and 4-finger arrays and observed a much higher success rate with 4-finger arrays. A variant of modular assembly that takes the context of neighboring fingers into account has also been reported and this method tends to yield proteins with improved performance relative to standard modular assembly.
Selection methods
Numerous selection methods have been used to generate zinc finger arrays capable of targeting desired sequences. Initial selection efforts utilized phage displayPhage display
Phage display is a method for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. Phage Display was originally invented by George P...
to select proteins that bound a given DNA target from a large pool of partially randomized zinc finger arrays. This technique is difficult to use on more than a single zinc finger at a time, so a multi-step processes that generated a completely optimized 3-finger array by adding and optimizing a single zinc finger at a time was developed.
More recent efforts have utilized yeast one-hybrid systems, bacterial one-hybrid and two-hybrid systems, and mammalian cells. A promising new method to select novel 3-finger zinc finger arrays utilizes a bacterial two-hybrid system and has been dubbed "OPEN" by its creators. This system combines pre-selected pools of individual zinc fingers that were each selected to bind a given triplet and then utilizes a second round of selection to obtain 3-finger arrays capable of binding a desired 9-bp sequence. This system was developed by the Zinc Finger Consortium as an alternative to commercial sources of engineered zinc finger arrays. It is somewhat difficult to directly compare the binding properties of proteins generated with this method to proteins generated by modular assembly as the specificity profiles of proteins generated by the OPEN method have never been reported.
Applications
Engineered zinc finger arrays can then be used in numerous applications such as artificial transcription factors, zinc finger methylases, zinc finger recombinases, and Zinc finger nucleases.While initial studies with another DNA-binding domain
DNA-binding domain
A DNA-binding domain is an independently folded protein domain that contains at least one motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence or have a general affinity to DNA...
from bacterial TAL effector
TAL effector
TAL effectors are proteins secreted by Xanthomonas bacteria via their type III secretion system when they infect various plant species...
s show promise, it remains to be seen whether these domains are suitable for some or all of the applications where engineered zinc fingers are currently used. Artificial transcription factors with engineered zinc finger arrays have been used in numerous scientific studies, and an artificial transcription factor that activates expression of VEGF
Vascular endothelial growth factor
Vascular endothelial growth factor is a signal protein produced by cells that stimulates vasculogenesis and angiogenesis. It is part of the system that restores the oxygen supply to tissues when blood circulation is inadequate....
is currently being evaluated in humans as a potential treatment for several clinical indications. Zinc finger nucleases have become useful reagents for manipulating genomes of many higher organisms including Drosophila melanogaster
Drosophila melanogaster
Drosophila melanogaster is a species of Diptera, or the order of flies, in the family Drosophilidae. The species is known generally as the common fruit fly or vinegar fly. Starting from Charles W...
, Caenorhabditis elegans
Caenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode , about 1 mm in length, which lives in temperate soil environments. Research into the molecular and developmental biology of C. elegans was begun in 1974 by Sydney Brenner and it has since been used extensively as a model...
, tobacco
Tobacco
Tobacco is an agricultural product processed from the leaves of plants in the genus Nicotiana. It can be consumed, used as a pesticide and, in the form of nicotine tartrate, used in some medicines...
, corn
Maize
Maize known in many English-speaking countries as corn or mielie/mealie, is a grain domesticated by indigenous peoples in Mesoamerica in prehistoric times. The leafy stalk produces ears which contain seeds called kernels. Though technically a grain, maize kernels are used in cooking as a vegetable...
,
zebrafish,
various types of mammalian cells,
and rats
RATS
RATS may refer to:* RATS , Regression Analysis of Time Series, a statistical package* Rough Auditing Tool for Security, a computer program...
.
An ongoing clinical trial is evaluating Zinc finger nucleases that disrupt the CCR5 gene in CD4+ human T-cells as a potential treatment for HIV/AIDS.
See also
- Zinc finger nucleaseZinc finger nucleaseZinc-finger nucleases are artificial restriction enzymes generated by fusing a zinc finger DNA-binding domain to a DNA-cleavage domain. Zinc finger domains can be engineered to target desired DNA sequences and this enables zinc-finger nucleases to target unique sequences within complex genomes...
- Zinc finger inhibitorZinc finger inhibitorZinc finger inhibition is the process by which the synthesis of zinc fingers is blocked. Zinc finger inhibitors have been tested for their efficacy in treating AIDS and HIV....
- Steroid hormone receptorSteroid hormone receptorSteroid hormone receptors are found on the plasma membrane, in the cytosol and also in the nucleus of target cells. They are generally intracellular receptors and initiate signal transduction for steroid hormones which lead to changes in gene expression over a time period of hours to days...
- DNA-binding proteinDNA-binding proteinDNA-binding proteins are proteins that are composed of DNA-binding domains and thus have a specific or general affinity for either single or double stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that...
- Sequence motifSequence motifIn genetics, a sequence motif is a nucleotide or amino-acid sequence pattern that is widespread and has, or is conjectured to have, a biological significance...
- Structural motifStructural motifIn a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a supersecondary structure, which appears also in a variety of other molecules...
- RING finger domainRING finger domainIn molecular biology, a RING finger domain is a protein structural domain of zinc finger type which contains a Cys3HisCys4 amino acid motif which binds two zinc cations. This protein domain contains from 40 to 60 amino acids...
- B-box zinc fingerB-box zinc fingerIn molecular biology the B-box-type zinc finger domain is a short protein domain of around 40 amino acid residues in length. B-box zinc fingers can be divided into two groups, where types 1 and 2 B-box domains differ in their consensus sequence and in the spacing of the 7-8 zinc-binding residues...
External links
- The double helix between the zinc finger
- Zinc Finger Tools design and information site
- Human KZNF Gene Catalog
- Zinc finger C2H2-type domain in PROSITEPROSITEPROSITE is a protein database. It consists of entries describing the protein families, domains and functional sites as well as amino acid patterns, signatures, and profiles in them. These are manually curated by a team of the Swiss Institute of Bioinformatics and tightly integrated into Swiss-Prot...
- Entry for zinc finger class C2H2 in the SMART database
- The Zinc Finger Consortium
- ZiFiT- Zinc Finger Design Tool
- Zinc Finger Consortium Materials from Addgene

