Transmembrane protein
Encyclopedia
A transmembrane protein is a protein that goes from one side of a membrane through to the other side of the membrane. Many TPs function as gateways or "loading docks" to deny or permit the transport of specific substances across the biological membrane, to get into the cell, or out of the cell as in the case of waste byproducts. As a response to the shape of certain molecules these "freight handling" TPs may have special ways of folding up or bending that will move a substance through the biological membrane.
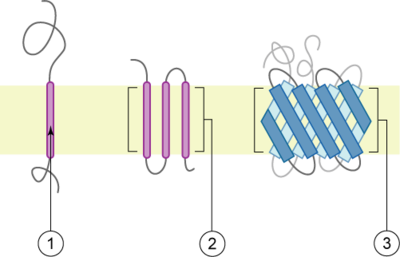
A transmembrane protein is a polytopic protein
that spans an entire biological membrane
. Transmembrane proteins aggregate and precipitate in water. They require detergent
s or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.
 All transmembrane proteins are integral membrane protein
All transmembrane proteins are integral membrane protein
s, but not all IMPs are transmembrane proteins.
Another classification refers to the position of the N- and C-terminal domains. Types I, II, and III are single pass molecules, while type IV are multiple pass molecules. Type I transmembrane proteins are anchored to the lipid membrane with a stop-transfer anchor sequence and have their N-terminal domains targeted to the ER lumen during synthesis (and the extracellular space, if mature forms are located on plasmalemma). Type II and III are anchored with a signal-anchor sequence, with type II being targeted to the ER lumen with its C-terminal domain, while type III have their N-terminal domains targeted to the ER lumen. Type IV is subdivided into IV-A, with their N-terminal domains targeted to the cytosol and IV-B, with an N-terminal domain targeted to the lumen. The implications for the division in the four types are especially manifest at the time of translocation and ER-bound translation, when the protein has to be passed through the ER membrane in a direction dependent on the type.
proteins are unusually stable judging from thermal denaturation
studies, because they do not unfold completely within the membranes (the complete unfolding would require breaking down too many α-helical H-bonds in the nonpolar media). On the other hand, these proteins easily misfold, due to non-native aggregation in membranes, transition to the molten globule
states, formation of non-native disulfide bonds, or unfolding of peripheral regions and nonregular loops that are locally less stable.
It is also important to properly define the unfolded state. The unfolded state of membrane proteins in detergent
micelles is different from that in the thermal denaturation
experiments. This state represents a combination of folded hydrophobic α-helices and partially unfolded segments covered by the detergent. For example, the "unfolded" bacteriorhodopsin
in SDS
micelles has four transmembrane α-helices folded, while the rest of the protein is situated at the micelle-water interface and can adopt different types of non-native amphiphilic structures. Free energy differences between such detergent-denatured and native states are similar to stabilities of water-soluble proteins (< 10 kcal/mol).
. In vivo all such proteins are normally folded co-translationally within the large transmembrane translocon
.The translocon channel provides a highly heterogeneous environment for the nascent transmembane α-helices. A relatively polar amphiphilic α-helix can adopt a transmembrane orientation in the translocon (although it would be at the membrane surface or unfolded in vitro), because its polar residues can face the central water-filled channel of the translocon. Such mechanism is necessary for incorporation of polar α-helices into structures of transmembrane proteins. The amphiphilic helices remain attached to the translocon until the protein is completely synthesized and folded. If the protein remains unfolded and attached to the translocon for too long, it is degraded by specific "quality control" cellular systems.
Note: n and S are, respectively, the number of beta-strands and the "shear number" of the beta-barrel
See also Gramicidin
A http://opm.phar.umich.edu/protein.php?pdbid=1grm, a peptide that forms a dimeric transmembrane β-helix. It is also secreted by Gram-positive
bacteria.
Transporter Classification database
A transmembrane protein is a polytopic protein
Protein
Proteins are biochemical compounds consisting of one or more polypeptides typically folded into a globular or fibrous form, facilitating a biological function. A polypeptide is a single linear polymer chain of amino acids bonded together by peptide bonds between the carboxyl and amino groups of...
that spans an entire biological membrane
Biological membrane
A biological membrane or biomembrane is an enclosing or separatingmembrane that acts as a selective barrier, within or around a cell. It consists of a lipid bilayer with embedded proteins that may constitute close to 50% of membrane content...
. Transmembrane proteins aggregate and precipitate in water. They require detergent
Detergent
A detergent is a surfactant or a mixture of surfactants with "cleaning properties in dilute solutions." In common usage, "detergent" refers to alkylbenzenesulfonates, a family of compounds that are similar to soap but are less affected by hard water...
s or nonpolar solvents for extraction, although some of them (beta-barrels) can be also extracted using denaturing agents.

Integral membrane protein
An integral membrane protein is a protein molecule that is permanently attached to the biological membrane. Proteins that cross the membrane are surrounded by "annular" lipids, which are defined as lipids that are in direct contact with a membrane protein...
s, but not all IMPs are transmembrane proteins.
Types
There are two basic types of transmembrane proteins:- Alpha-helical. These proteins are present in the inner membranes of bacterial cells or the plasma membrane of eukaryotes, and sometimes in the outer membraneOuter membraneThe bacterial outer membrane is found in Gram-negative bacteria. Its composition is distinct from that of the cytoplasmic membrane - among other things, the outer leaflet of the membrane includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and it is linked to the cell's...
s. This is the major category of transmembrane proteins. In humans, 27% of all proteins have been estimated to be alpha-helical membrane proteins. - Beta-barrels. These proteins are so far found only in outer membraneOuter membraneThe bacterial outer membrane is found in Gram-negative bacteria. Its composition is distinct from that of the cytoplasmic membrane - among other things, the outer leaflet of the membrane includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and it is linked to the cell's...
s of Gram-negative bacteria, cell wallCell wallThe cell wall is the tough, usually flexible but sometimes fairly rigid layer that surrounds some types of cells. It is located outside the cell membrane and provides these cells with structural support and protection, and also acts as a filtering mechanism. A major function of the cell wall is to...
of Gram-positive bacteria, and outer membraneOuter membraneThe bacterial outer membrane is found in Gram-negative bacteria. Its composition is distinct from that of the cytoplasmic membrane - among other things, the outer leaflet of the membrane includes a complex lipopolysaccharide whose lipid portion acts as an endotoxin - and it is linked to the cell's...
s of mitochondria and chloroplasts. All beta-barrel transmembrane proteins have simplest up-and-down topology, which may reflect their common evolutionary origin and similar folding mechanism.
Another classification refers to the position of the N- and C-terminal domains. Types I, II, and III are single pass molecules, while type IV are multiple pass molecules. Type I transmembrane proteins are anchored to the lipid membrane with a stop-transfer anchor sequence and have their N-terminal domains targeted to the ER lumen during synthesis (and the extracellular space, if mature forms are located on plasmalemma). Type II and III are anchored with a signal-anchor sequence, with type II being targeted to the ER lumen with its C-terminal domain, while type III have their N-terminal domains targeted to the ER lumen. Type IV is subdivided into IV-A, with their N-terminal domains targeted to the cytosol and IV-B, with an N-terminal domain targeted to the lumen. The implications for the division in the four types are especially manifest at the time of translocation and ER-bound translation, when the protein has to be passed through the ER membrane in a direction dependent on the type.
Stability of α-helical transmembrane proteins
Transmembrane α-helicalTransmembrane helix
Transmembrane domain usually denotes a single transmembrane alpha helix of a transmembrane protein. It is called a "domain" because an alpha-helix in a membrane can fold independently from the rest of the protein, similar to domains of water-soluble proteins...
proteins are unusually stable judging from thermal denaturation
Denaturation (biochemistry)
Denaturation is a process in which proteins or nucleic acids lose their tertiary structure and secondary structure by application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent , or heat...
studies, because they do not unfold completely within the membranes (the complete unfolding would require breaking down too many α-helical H-bonds in the nonpolar media). On the other hand, these proteins easily misfold, due to non-native aggregation in membranes, transition to the molten globule
Molten globule
The term molten globule was first coined by A. Wada and M Ohgushi in 1983. It was first found in cytochrome c, which conserves a native-like secondary structure content but without the tightly packed protein interior, under low pH and high salt concentration...
states, formation of non-native disulfide bonds, or unfolding of peripheral regions and nonregular loops that are locally less stable.
It is also important to properly define the unfolded state. The unfolded state of membrane proteins in detergent
Detergent
A detergent is a surfactant or a mixture of surfactants with "cleaning properties in dilute solutions." In common usage, "detergent" refers to alkylbenzenesulfonates, a family of compounds that are similar to soap but are less affected by hard water...
micelles is different from that in the thermal denaturation
Denaturation (biochemistry)
Denaturation is a process in which proteins or nucleic acids lose their tertiary structure and secondary structure by application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent , or heat...
experiments. This state represents a combination of folded hydrophobic α-helices and partially unfolded segments covered by the detergent. For example, the "unfolded" bacteriorhodopsin
Bacteriorhodopsin
Bacteriorhodopsin is a protein used by Archaea, the most notable one being Halobacteria. It acts as a proton pump; that is, it captures light energy and uses it to move protons across the membrane out of the cell...
in SDS
Sodium dodecyl sulfate
Sodium dodecyl sulfate , sodium laurilsulfate or sodium lauryl sulfate is an organic compound with the formula CH311OSO3Na). It is an anionic surfactant used in many cleaning and hygiene products...
micelles has four transmembrane α-helices folded, while the rest of the protein is situated at the micelle-water interface and can adopt different types of non-native amphiphilic structures. Free energy differences between such detergent-denatured and native states are similar to stabilities of water-soluble proteins (< 10 kcal/mol).
Folding of α-helical transmembrane proteins
Refolding of α-helical transmembrane proteins in vitro is technically difficult. There are relatively few examples of the successful refolding experiments, as for bacteriorhodopsinBacteriorhodopsin
Bacteriorhodopsin is a protein used by Archaea, the most notable one being Halobacteria. It acts as a proton pump; that is, it captures light energy and uses it to move protons across the membrane out of the cell...
. In vivo all such proteins are normally folded co-translationally within the large transmembrane translocon
Translocon
The translocon is the complex of proteins associated with the translocation of nascent polypeptides across membranes. In eukaryotes the polypeptides are transported into the interior space of the endoplasmic reticulum from the cytosol...
.The translocon channel provides a highly heterogeneous environment for the nascent transmembane α-helices. A relatively polar amphiphilic α-helix can adopt a transmembrane orientation in the translocon (although it would be at the membrane surface or unfolded in vitro), because its polar residues can face the central water-filled channel of the translocon. Such mechanism is necessary for incorporation of polar α-helices into structures of transmembrane proteins. The amphiphilic helices remain attached to the translocon until the protein is completely synthesized and folded. If the protein remains unfolded and attached to the translocon for too long, it is degraded by specific "quality control" cellular systems.
Stability and folding of β-barrel transmembrane proteins
Stability of β-barrel transmembrane proteins is similar to stability of water-soluble proteins, based on chemical denaturation studies. Their folding in vivo is facilitated by water-soluble chaperones, such as protein Skp http://faculty.virginia.edu/tamm/pages/BBA_Folding_Review.pdf.Light absorption-driven transporters
- BacteriorhodopsinBacteriorhodopsinBacteriorhodopsin is a protein used by Archaea, the most notable one being Halobacteria. It acts as a proton pump; that is, it captures light energy and uses it to move protons across the membrane out of the cell...
-like proteins including rhodopsinRhodopsinRhodopsin, also known as visual purple, is a biological pigment of the retina that is responsible for both the formation of the photoreceptor cells and the first events in the perception of light. Rhodopsins belong to the G-protein coupled receptor family and are extremely sensitive to light,...
(see also opsinOpsinOpsins are a group of light-sensitive 35–55 kDa membrane-bound G protein-coupled receptors of the retinylidene protein family found in photoreceptor cells of the retina. Five classical groups of opsins are involved in vision, mediating the conversion of a photon of light into an electrochemical...
)http://opm.phar.umich.edu/families.php?superfamily=6 - Bacterial photosynthetic reaction centrePhotosynthetic reaction centreA photosynthetic reaction center is a complex of several proteins, pigments and other co-factors assembled together to execute the primary energy conversion reactions of photosynthesis...
s and photosystemPhotosystemPhotosystems are functional and structural units of protein complexes involved in photosynthesis that together carry out the primary photochemistry of photosynthesis: the absorption of light and the transfer of energy and electrons...
s I and II http://opm.phar.umich.edu/families.php?superfamily=2 - Light harvesting complexes from bacteriaBacteriaBacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
and chloroplasts http://opm.phar.umich.edu/families.php?superfamily=1
Oxidoreduction-driven transporters
- Transmembrane cytochrome b-like proteins http://opm.phar.umich.edu/families.php?superfamily=3: coenzyme Q - cytochrome c reductaseCoenzyme Q - cytochrome c reductaseIn enzymology, a ubiquinol—cytochrome-c reductase is an enzyme that catalyzes the chemical reactionThus, the two substrates of this enzyme are dihydroquinone and ferri- cytochrome c, whereas its 3 products are quinone , ferro- cytochrome c, and H+.This enzyme belongs to the family of...
(cytochrome bc1 ); cytochrome b6f complexCytochrome b6f complexThe cytochrome b6f complex is an enzyme found in the thylakoid membrane in chloroplasts of plants, cyanobacteria, and green algae, catalyzing the transfer of electrons from plastoquinol to plastocyanin...
; formate dehydrogenase, respiratory nitrate reductaseNitrate reductaseNitrate reductases are molybdoenzymes that reduce nitrate to nitrite .* Eukaryotic nitrate reductases are part of the sulfite oxidase family of molybdoenzymes....
; succinate - coenzyme Q reductaseSuccinate - coenzyme Q reductaseSuccinate dehydrogenase or succinate-coenzyme Q reductase or Complex II is an enzyme complex, bound to the inner mitochondrial membrane of mammalian mitochondria and many bacterial cells...
(fumarate reductase); and succinate dehydrogenase. See electron transport chainElectron transport chainAn electron transport chain couples electron transfer between an electron donor and an electron acceptor with the transfer of H+ ions across a membrane. The resulting electrochemical proton gradient is used to generate chemical energy in the form of adenosine triphosphate...
. - Cytochrome c oxidaseCytochrome c oxidaseThe enzyme cytochrome c oxidase or Complex IV is a large transmembrane protein complex found in bacteria and the mitochondrion.It is the last enzyme in the respiratory electron transport chain of mitochondria located in the mitochondrial membrane...
s http://opm.phar.umich.edu/families.php?superfamily=4 from bacteriaBacteriaBacteria are a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria have a wide range of shapes, ranging from spheres to rods and spirals...
and mitochondria
Electrochemical potential-driven transporters
- Proton or sodium translocating F-type and V-type ATPaseATPaseATPases are a class of enzymes that catalyze the decomposition of adenosine triphosphate into adenosine diphosphate and a free phosphate ion. This dephosphorylation reaction releases energy, which the enzyme harnesses to drive other chemical reactions that would not otherwise occur...
s http://opm.phar.umich.edu/families.php?superfamily=5
P-P-bond hydrolysis-driven transporters
- P-type calcium ATPaseCalcium ATPaseCalcium ATPase is a form of P-ATPase that transfers calcium after a muscle has contracted. The calcium ATPase are:*Plasma membrane Ca2+ ATPase *Sarcoplasmic reticulum Ca2+ ATPase - Plasma membrane Ca2+ ATPase :...
(five different conformations) http://opm.phar.umich.edu/families.php?superfamily=22 - Calcium ATPase regulators phospholambanPhospholambanPhospholamban, also known as PLN, is a protein that in humans is encoded by the PLN gene. Phospholamban is a 52-amino acid integral membrane protein that regulates the Ca2+ pump in cardiac muscle and skeletal muscle cells.- Function :...
and sarcolipin http://opm.phar.umich.edu/families.php?superfamily=70 - ABC transporters: BtuCD, multidrug transporter, and molybdate uptake transporter
- General secretory pathwaySecretory pathwayThe secretory pathway is a series of steps a cell uses to move proteins out of the cell; a process known as secretion. The path of a protein destined for secretion has its origins in the rough endoplasmic reticulum, a membrane-bound compartment in the cell...
(Sec) transloconTransloconThe translocon is the complex of proteins associated with the translocation of nascent polypeptides across membranes. In eukaryotes the polypeptides are transported into the interior space of the endoplasmic reticulum from the cytosol...
(preprotein translocase SecY) http://opm.phar.umich.edu/families.php?superfamily=19
Porters (uniporters, symporters, antiporters)
- Mitochondrial carrier proteinCarrier proteinCarrier proteins are proteins involved in the movement of ions, small molecules, or macromolecules, such as another protein, across a biological membrane. Carrier proteins are integral membrane proteins; that is they exist within and span the membrane across which they transport substances. The...
s http://opm.phar.umich.edu/families.php?superfamily=21 - Major Facilitator Superfamily (Glycerol-3-phosphate transporter, Lactose permeasePermeaseThe permeases are membrane transport proteins, a class of multipass transmembrane proteins that facilitate the diffusion of a specific molecule in or out of the cell by passive transport...
, and Multidrug transporter EmrD) http://opm.phar.umich.edu/families.php?superfamily=15 - Resistance-nodulation-cell division (multidrug efflux transporter AcrB, see multidrug resistanceMultidrug resistanceMultiple drug resistance or Multidrug resistance is a condition enabling a disease-causing organism to resist distinct drugs or chemicals of a wide variety of structure and function targeted at eradicating the organism...
)http://opm.phar.umich.edu/families.php?superfamily=16 - Dicarboxylate/amino acid:cation symporter (proton glutamate symporter) http://opm.phar.umich.edu/families.php?superfamily=20
- Monovalent cation/proton antiporter (Sodium/proton antiporter 1 NhaA) http://opm.phar.umich.edu/families.php?superfamily=66
- NeurotransmitterNeurotransmitterNeurotransmitters are endogenous chemicals that transmit signals from a neuron to a target cell across a synapse. Neurotransmitters are packaged into synaptic vesicles clustered beneath the membrane on the presynaptic side of a synapse, and are released into the synaptic cleft, where they bind to...
sodium symporter http://opm.phar.umich.edu/families.php?superfamily=67 - Ammonia transporters http://opm.phar.umich.edu/families.php?superfamily=13
- Drug/Metabolite Transporter (small multidrug resistance transporter EmrE - the structures are retracted as erroneous) http://opm.phar.umich.edu/families.php?superfamily=77
Alpha-helical channels including ion channels
- Voltage-gated ion channelVoltage-gated ion channelVoltage-gated ion channels are a class of transmembrane ion channels that are activated by changes in electrical potential difference near the channel; these types of ion channels are especially critical in neurons, but are common in many types of cells....
like, including potassium channelPotassium channelIn the field of cell biology, potassium channels are the most widely distributed type of ion channel and are found in virtually all living organisms. They form potassium-selective pores that span cell membranes...
s KcsA and KvAP, and inward-rectifier potassium ion channelInward-rectifier potassium ion channelInwardly rectifying potassium channels are a specific subset of potassium selective ion channels. To date, seven subfamilies have been identified in various mammalian cell types...
Kirbac http://opm.phar.umich.edu/families.php?superfamily=8 - Large-conductance mechanosensitive channel, MscL http://opm.phar.umich.edu/families.php?superfamily=12
- Small-conductance mechanosensitive ion channel (MscS)Mechanosensitive ion channelMechanosensitive channels are found in a number of tissues and organisms and are thought to be the sensors for a number of systems including the senses of touch, hearing and balance, as well as participating in cardiovascular regulation and osmotic homeostasis...
http://opm.phar.umich.edu/families.php?superfamily=11 - CorA metal ion transporters http://opm.phar.umich.edu/families.php?superfamily=72
- Ligand-gated ion channelLigand-gated ion channelLigand-gated ion channels are one type of ionotropic receptor or channel-linked receptor. They are a group of transmembrane ion channels that are opened or closed in response to the binding of a chemical messenger , such as a neurotransmitter.The binding site of endogenous ligands on LGICs...
of neurotransmitterNeurotransmitterNeurotransmitters are endogenous chemicals that transmit signals from a neuron to a target cell across a synapse. Neurotransmitters are packaged into synaptic vesicles clustered beneath the membrane on the presynaptic side of a synapse, and are released into the synaptic cleft, where they bind to...
receptors (acetylcholine receptorAcetylcholine receptorAn acetylcholine receptor is an integral membrane protein that responds to the binding of acetylcholine, a neurotransmitter.-Classification:...
) http://opm.phar.umich.edu/families.php?superfamily=14 - AquaporinAquaporinAquaporins are proteins embedded in the cell membrane that regulate the flow of water.Aquaporins are integral membrane proteins from a larger family of major intrinsic proteins that form pores in the membrane of biological cells....
s http://opm.phar.umich.edu/families.php?superfamily=7 - Chloride channelChloride channelChloride channels are a superfamily of poorly understood ion channels consisting of approximately 13 members.Chloride channels display a variety of important physiological and cellular roles that include regulation of pH, volume homeostasis, organic solute transport, cell migration, cell...
s http://opm.phar.umich.edu/families.php?superfamily=10 - Outer membrane auxiliary proteins (polysaccharide transporter) http://opm.phar.umich.edu/families.php?superfamily=188 - α-helical transmembrane proteins from the outer bacterial membrane
Enzymes
- Methane monooxygenaseMethane monooxygenaseMethane monooxygenase, or MMO, is an enzyme capable of oxidizing the C-H bond in methane as well as other alkanes. Methane monooxygenase belongs to the class of oxidoreductase enzymes ....
http://opm.phar.umich.edu/families.php?superfamily=23 - Rhomboid protease http://opm.phar.umich.edu/families.php?superfamily=186
- Disulfide bondDisulfide bondIn chemistry, a disulfide bond is a covalent bond, usually derived by the coupling of two thiol groups. The linkage is also called an SS-bond or disulfide bridge. The overall connectivity is therefore R-S-S-R. The terminology is widely used in biochemistry...
formation protein (DsbA-DsbB complex) http://opm.phar.umich.edu/protein.php?pdbid=2hi7
Proteins with alpha-helical transmembrane anchors
- T cell receptorT cell receptorThe T cell receptor or TCR is a molecule found on the surface of T lymphocytes that is responsible for recognizing antigens bound to major histocompatibility complex molecules...
transmembrane dimerization domain http://opm.phar.umich.edu/families.php?superfamily=187 - Cytochrome c nitrite reductaseNitrite reductaseNitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2 to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.- Iron based...
complex http://opm.phar.umich.edu/protein.php?pdbid=2j7a - Steryl-sulfate sulfohydrolase http://opm.phar.umich.edu/families.php?superfamily=24
- Stannin http://opm.phar.umich.edu/families.php?superfamily=180
- GlycophorinGlycophorinA Glycophorin is a sialoglycoprotein of the membrane of a red blood cell. It is a membrane-spanning protein and carries sugar molecules. It is heavily glycosylated . Glycophorins are rich in sialic acid, which gives the red cells a very hydrophilic-charged coat...
A dimer http://opm.phar.umich.edu/families.php?superfamily=25 - Inovirus (filamentous phageFilamentous phageA filamentous phage is a type of bacteriophage shaped like a rod filament. Filamentous phages usually contain a genome of single-stranded DNA and infect Gram-negative bacteria.-Types of filamentous phage:*Ff phages - these infect E...
) major coat protein http://opm.phar.umich.edu/families.php?superfamily=73 - PilinPilinPilin refers to a class of fibrous proteins that are found in pilus structures in bacteria. Bacterial pili are used in the exchange of genetic material during bacterial conjugation, and a short pilus called a fimbrium is used as a cell adhesion mechanism. Although not all bacteria have pili or...
http://opm.phar.umich.edu/families.php?superfamily=74 - Pulmonary surfactantPulmonary surfactantPulmonary surfactant is a surface-active lipoprotein complex formed by type II alveolar cells. The proteins and lipids that surfactant comprises have both a hydrophilic region and a hydrophobic region...
-associated protein http://opm.phar.umich.edu/families.php?superfamily=75 - Monoamine oxidaseMonoamine oxidaseL-Monoamine oxidases are a family of enzymes that catalyze the oxidation of monoamines. They are found bound to the outer membrane of mitochondria in most cell types in the body. The enzyme was originally discovered by Mary Bernheim in the liver and was named tyramine oxidase...
s A and B http://opm.phar.umich.edu/families.php?family=176, - Fatty acid amide hydrolase
- Cytochrome P450 oxidaseCytochrome P450 oxidaseThe cytochrome P450 superfamily is a large and diverse group of enzymes. The function of most CYP enzymes is to catalyze the oxidation of organic substances. The substrates of CYP enzymes include metabolic intermediates such as lipids and steroidal hormones, as well as xenobiotic substances...
s http://opm.phar.umich.edu/families.php?superfamily=41, - Corticosteroid 11β-dehydrogenasesMineralocorticoidMineralocorticoids are a class of steroid hormones characterised by their similarity to aldosterone and their influence on salt and water balances.-Physiology:...
http://opm.phar.umich.edu/families.php?superfamily=127. - Signal Peptide PeptidaseSignal Peptide PeptidaseThe Signal Peptide Peptidase is an intramembrane aspartyl protease with the conserved active site motifs 'YD' and 'GxGD' in in adjacent transmembrane domains . Its sequences is highly conserved in different vertebrate species...
http://opm.phar.umich.edu/families.php?superfamily=178 - Membrane protease specific for a stomatin homolog http://opm.phar.umich.edu/protein.php?pdbid=2deo
β-barrels composed of a single polypeptide chain
- Beta barrels from eight beta-strands and with "shear number" of ten (n=8, S=10) http://opm.phar.umich.edu/families.php?superfamily=26. They include:
- OmpA-like transmembrane domainOmpA-like transmembrane domainOmpA-like transmembrane domain is an evolutionarily conserved domain of outer membrane proteins. This domain consists of an eight-stranded beta barrel. OmpA is the predominant cell surface antigen in enterobacteria found in about 100,000 copies per cell. The expression of OmpA is tightly regulated...
(OmpA), - Virulence-related outer membrane protein familyVirulence-related outer membrane protein familyVirulence-related outer membrane proteins are expressed in Gram-negative bacteria and are essential to bacterial survival within macrophages and for eukaryotic cell invasion.This family consists of several bacterial and phage Ail/Lom-like proteins....
(OmpX), - Outer membrane protein W familyOuter membrane protein W familyOuter membrane protein W family is a family of evolutionarily related proteins from the outer bacterial membrane.This family includes outer membrane protein W proteins from a variety of bacterial species. This protein may form the receptor for S4 colicins in Escherichia coli....
(OmpW), - Antimicrobial peptide resistance and lipid A acylation protein familyAntimicrobial peptide resistance and lipid A acylation protein familyAntimicrobial peptide resistance and lipid A acylation protein PagP is a family of several bacterial antimicrobial peptide resistance and lipid A acylation proteins. The bacterial outer membrane enzyme PagP transfers a palmitate chain from a phospholipid to lipid A...
(PagP) - Lipid A deacylase PagLLipid A deacylase PagLLipid A deacylase is an outer membrane protein with lipid A 3-O-deacylase activity. It forms an 8 stranded beta barrel structure.-References:* Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa...
, and - Opacity family porinsOpacity family porinsOpacity family porins are a family of porins from pathogenic Neisseria.These bacteria possess a repertoire of phase-variable opacity proteins that mediate various pathogen/host cell interactions. These proteins are related to OmpA-like transmembrane domain family....
(NspA)
- OmpA-like transmembrane domain
- Autotransporter domainAutotransporter domainIn molecular biology an autotransporter domain is a structural domain found in some outer membrane proteins.Translocation of polypeptide chains through the outer membrane of Gram-negative bacteria is termed secretion. Secretion occurs via a number of different pathways in this type of bacterium...
(n=12,S=14') http://opm.phar.umich.edu/families.php?superfamily=28 - FadL outer membrane protein transport familyFadL outer membrane protein transport familyOuter membrane transport proteins family includes several proteins that are involved in toluene catabolism and degradation of aromatic hydrocarbons. This family also includes protein FadL involved in translocation of long-chain...
, including Fatty acidFatty acidIn chemistry, especially biochemistry, a fatty acid is a carboxylic acid with a long unbranched aliphatic tail , which is either saturated or unsaturated. Most naturally occurring fatty acids have a chain of an even number of carbon atoms, from 4 to 28. Fatty acids are usually derived from...
transporter FadL (n=14,S=14) http://opm.phar.umich.edu/families.php?superfamily=30 - General bacterial porin familyGeneral bacterial porin familyGeneral bacterial porins are a family of proteins from the outer membrane of Gram-negative bacteria. The porins act as molecular filters for hydrophilic compounds. They are responsible for the 'molecular sieve' properties of the outer membrane. Porins form large water-filled channels which allow...
, known as trimeric porinsPorin (protein)Porins are beta barrel proteins that cross a cellular membrane and act as a pore through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules...
(n=16,S=20) http://opm.phar.umich.edu/families.php?superfamily=31 - MaltoporinMaltoporinMaltoporins are a family of outer membrane proteins. Maltoporin forms a trimeric structure which facilitates the diffusion of maltodextrins across the outer membrane of Gram-negative bacteria. The membrane channel is formed by an antiparallel beta-barrel....
, or sugar porinsPorin (protein)Porins are beta barrel proteins that cross a cellular membrane and act as a pore through which molecules can diffuse. Unlike other membrane transport proteins, porins are large enough to allow passive diffusion, i.e., they act as channels that are specific to different types of molecules...
(n=18,S=22) http://opm.phar.umich.edu/families.php?superfamily=32 - Nucleoside-specific porinNucleoside-specific porinNucleoside-specific porin is an outer membrane protein, Tsx, which constitutes the receptor for colicin K and Bacteriophage T6, and functions as a substrate-specific channel for nucleosides and deoxy-nucleosides. The protein contains 294 amino acids, the first 22 of which are characteristic of a...
(n=12,S=16) http://opm.phar.umich.edu/families.php?superfamily=71 - Outer membrane phospholipase A1Outer membrane phospholipase A1Outer membrane phospholipase A1 is an acyl hydrolasewith a broad substrate specificity from the bacterial outer membrane. It has been proposed that Ser164 is the active site of the protein...
(n=12,S=16) http://opm.phar.umich.edu/families.php?superfamily=29 - TonB-dependent receptorsTonB-dependent receptorsTonB-dependent receptors is a family of beta-barrel proteins from the outer membrane of Gram-negative bacteria. The TonB complex senses signals from outside the bacterial cell and transmits them via two membranes into the cytoplasm, leading to transcriptional activation of target genes.In...
and their plug domain. They are ligand-gated outer membrane channels (n=22,S=24), including cobalamin transporter BtuB, Fe(III)-pyochelin receptor FptA, receptor FepA, ferric hydroxamate uptake receptor FhuA, transporter FecA, and pyoverdine receptor FpvA http://opm.phar.umich.edu/families.php?superfamily=33 - Outer membrane protein OpcAOuter membrane protein OpcAOuter membrane adhesin OpcA protein family consists of several Neisseria species specific outer membrane proteins. Neisseria meningitidis causes meningococcal meningitis and septicemia...
family (n=10,S=12) that includes outer membrane proteaseProteaseA protease is any enzyme that conducts proteolysis, that is, begins protein catabolism by hydrolysis of the peptide bonds that link amino acids together in the polypeptide chain forming the protein....
OmpT and adhesinAdhesinAdhesins are cell-surface components or appendages of bacteria that facilitate bacterial adhesion or adherence to other cells or to inanimate surfaces. Adhesins are a type of virulence factor....
/invasin OpcA protein http://opm.phar.umich.edu/families.php?superfamily=27 - Outer membrane protein GOuter membrane protein GOuter membrane protein G is a porin, a channel proteins in the outer membrane of Gram-negative bacteria.Escherichia coli OmpG forms a 14-stranded beta-barrel and in contrast to most porins, appears to function as a monomer. The central pore of OmpG is wider than other E...
porin family (n=14,S=16) http://opm.phar.umich.edu/families.php?superfamily=181
Note: n and S are, respectively, the number of beta-strands and the "shear number" of the beta-barrel
β-barrels composed of several polypeptide chains
- Trimeric autotransporter (n=12,S=12) http://opm.phar.umich.edu/families.php?superfamily=179
- Outer membrane efflux proteinsOuter membrane efflux proteinsOuter membrane efflux protein family form trimericchannels that allow export of a variety of substrates in Gramnegative bacteria. Each member of this family is composed oftwo repeats...
, also known as trimeric outer membrane factors (n=12,S=18) including TolC and multidrug resistance proteins http://opm.phar.umich.edu/families.php?superfamily=34 - MspA porinMspA porinMspA porin is a membrane porin produced by Mycobacteria, allowing hydrophilic nutrients to enter the bacterium. The protein forms a tightly interconnected octamer with eightfold rotation symmetry that resembles a goblet and contains a central channel. Each subunit fold contains a beta-sandwich of...
(octamer, n=S=16) and α-hemolysin (heptamer n=S=14) http://opm.phar.umich.edu/families.php?superfamily=35. These proteins are secreted.
See also Gramicidin
Gramicidin
Gramicidin is a heterogeneous mixture of six antibiotic compounds, gramicidins A, B and C, making up 80%, 6%,and 14% respectively, all of which are obtained from the soil bacterial species Bacillus brevis and called collectively gramicidin D. Gramicidin D are linear pentadecapeptides; that is...
A http://opm.phar.umich.edu/protein.php?pdbid=1grm, a peptide that forms a dimeric transmembrane β-helix. It is also secreted by Gram-positive
Gram-positive
Gram-positive bacteria are those that are stained dark blue or violet by Gram staining. This is in contrast to Gram-negative bacteria, which cannot retain the crystal violet stain, instead taking up the counterstain and appearing red or pink...
bacteria.
See also
- cell membraneCell membraneThe cell membrane or plasma membrane is a biological membrane that separates the interior of all cells from the outside environment. The cell membrane is selectively permeable to ions and organic molecules and controls the movement of substances in and out of cells. It basically protects the cell...
- transmembrane receptors
- membrane topologyMembrane topologyIn biochemistry, the membrane topology of an transmembrane protein describes which portions of the amino-acid sequence of the protein lie within the plane of the surrounding lipid bilayer and which portions protrude into the watery environment on either side...
- transmembrane helixTransmembrane helixTransmembrane domain usually denotes a single transmembrane alpha helix of a transmembrane protein. It is called a "domain" because an alpha-helix in a membrane can fold independently from the rest of the protein, similar to domains of water-soluble proteins...
- membrane proteinMembrane proteinA membrane protein is a protein molecule that is attached to, or associated with the membrane of a cell or an organelle. More than half of all proteins interact with membranes.-Function:...
- integral membrane proteinIntegral membrane proteinAn integral membrane protein is a protein molecule that is permanently attached to the biological membrane. Proteins that cross the membrane are surrounded by "annular" lipids, which are defined as lipids that are in direct contact with a membrane protein...
- peripheral membrane proteinPeripheral membrane proteinPeripheral membrane proteins are proteins that adhere only temporarily to the biological membrane with which they are associated. These molecules attach to integral membrane proteins, or penetrate the peripheral regions of the lipid bilayer. The regulatory protein subunits of many ion channels and...
- intramembrane proteaseIntramembrane proteaseIntramembrane proteases are enzymes that have the property of cleaving transmembrane domains of integral membrane proteins. All known intramembrane proteases are themselves integral membrane proteins with multiple transmembrane domains, and they have their active sites buried within the lipid...
Transporter Classification database

