
Inverted index
Encyclopedia
In computer science
, an inverted index (also referred to as postings file or inverted file) is an index data structure
storing a mapping from content, such as words or numbers, to its locations in a database file
, or in a document or a set of documents. The purpose of an inverted index is to allow fast full text search
es, at a cost of increased processing when a document is added to the database. The inverted file may be the database file itself, rather than its index
. It is the most popular data structure used in document retrieval
systems, used on a large scale for example in search engine
s. Several significant general-purpose mainframe
-based database management systems have used inverted list architectures, including ADABAS
, DATACOM/DB
, and Model 204
.
There are two main variants of inverted indexes: A record level inverted index (or inverted file index or just inverted file) contains a list of references to documents for each word. A word level inverted index (or full inverted index or inverted list) additionally contains the positions of each word within a document. The latter form offers more functionality (like phrase search
es), but needs more time and space to be created.

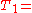
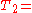
we have the following inverted file index (where the integers in the set notation brackets refer to the subscripts of the text symbols, ,
,  etc.):
etc.):
"a": {2}
"banana": {2}
"is": {0, 1, 2}
"it": {0, 1, 2}
"what": {0, 1}
A term search for the terms
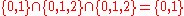 .
.
With the same texts, we get the following full inverted index, where the pairs are document numbers and local word numbers. Like the document numbers, local word numbers also begin with zero. So, ), and it is the fourth word in that document (position 3).
), and it is the fourth word in that document (position 3).
"a": {(2, 2)}
"banana": {(2, 3)}
"is": {(0, 1), (0, 4), (1, 1), (2, 1)}
"it": {(0, 0), (0, 3), (1, 2), (2, 0)}
"what": {(0, 2), (1, 0)}
If we run a phrase search for
is a central component of a typical search engine indexing algorithm
. A goal of a search engine implementation is to optimize the speed of the query: find the documents where word X occurs. Once a forward index is developed, which stores lists of words per document, it is next inverted to develop an inverted index. Querying the forward index would require sequential iteration through each document and to each word to verify a matching document. The time, memory, and processing resources to perform such a query are not always technically realistic. Instead of listing the words per document in the forward index, the inverted index data structure is developed which lists the documents per word.
With the inverted index created, the query can now be resolved by jumping to the word id (via random access
) in the inverted index.
In pre-computer times, concordances
to important books were manually assembled. These were effectively inverted indexes with a small amount of accompanying commentary, that required a tremendous amount of effort to produce.
In bioinformatics, inverted indexes are very important in the sequence assembly
of short fragments of sequenced DNA. One way to find out where fragments came from is to search for it against a reference DNA sequence. A small number of mismatches (due to differences between the sequenced DNA and reference DNA, or due to errors) can be accounted for by dividing the fragment into smaller fragments—at least one subfragment is likely to match the reference DNA sequence. The matching requires constructing an inverted index of all substrings of a certain length from the reference DNA sequence. Since the human DNA contains more than 3 billion base pairs, and we need to store a DNA substring for every index, and a 32-bit integer for index itself, the storage requirement for such an inverted index would probably be in the tens of gigabytes, just beyond the available RAM capacity of most personal computers today.
Computer science
Computer science or computing science is the study of the theoretical foundations of information and computation and of practical techniques for their implementation and application in computer systems...
, an inverted index (also referred to as postings file or inverted file) is an index data structure
Index (information technology)
In computer science, an index can be:# an integer that identifies an array element# a data structure that enables sublinear-time lookup -Array element identifier:...
storing a mapping from content, such as words or numbers, to its locations in a database file
Table (database)
In relational databases and flat file databases, a table is a set of data elements that is organized using a model of vertical columns and horizontal rows. A table has a specified number of columns, but can have any number of rows...
, or in a document or a set of documents. The purpose of an inverted index is to allow fast full text search
Full text search
In text retrieval, full text search refers to techniques for searching a single computer-stored document or a collection in a full text database...
es, at a cost of increased processing when a document is added to the database. The inverted file may be the database file itself, rather than its index
Index (database)
A database index is a data structure that improves the speed of data retrieval operations on a database table at the cost of slower writes and increased storage space...
. It is the most popular data structure used in document retrieval
Document retrieval
Document retrieval is defined as the matching of some stated user query against a set of free-text records. These records could be any type of mainly unstructured text, such as newspaper articles, real estate records or paragraphs in a manual...
systems, used on a large scale for example in search engine
Search engine
A search engine is an information retrieval system designed to help find information stored on a computer system. The search results are usually presented in a list and are commonly called hits. Search engines help to minimize the time required to find information and the amount of information...
s. Several significant general-purpose mainframe
Mainframe computer
Mainframes are powerful computers used primarily by corporate and governmental organizations for critical applications, bulk data processing such as census, industry and consumer statistics, enterprise resource planning, and financial transaction processing.The term originally referred to the...
-based database management systems have used inverted list architectures, including ADABAS
Adabas
ADABAS is Software AG’s primary database management system.- History :First released in 1970, ADABAS is considered by some to have been one of the earliest commercially available database products...
, DATACOM/DB
DATACOM/DB
Datacom/DB is a relational database management system for the mainframes. Originally developed by Insyte Datacom, later acquired by Applied Data Research, it is now owned by CA Technologies which renamed it to CA-Datacom/DB and later to CA Datacom/DB.- External links :****...
, and Model 204
Model 204
Model 204 is a Database management system for IBM and compatible mainframes, which was first deployed in 1972. It incorporates a programming language and an environment for application development. It can deal with very large databases and very high transaction loads.Model 204 relies on its own...
.
There are two main variants of inverted indexes: A record level inverted index (or inverted file index or just inverted file) contains a list of references to documents for each word. A word level inverted index (or full inverted index or inverted list) additionally contains the positions of each word within a document. The latter form offers more functionality (like phrase search
Phrase search
Phrase Search is a type of search that allows users to search for documents containing an exact sentence or phrase opposed to being limited to keywords...
es), but needs more time and space to be created.
Example
Given the texts
"it is what it is",
"what is it" and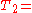
"it is a banana",we have the following inverted file index (where the integers in the set notation brackets refer to the subscripts of the text symbols,
 ,
,  etc.):
etc.):"a": {2}
"banana": {2}
"is": {0, 1, 2}
"it": {0, 1, 2}
"what": {0, 1}
A term search for the terms
"what", "is" and "it" would give the set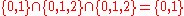 .
.With the same texts, we get the following full inverted index, where the pairs are document numbers and local word numbers. Like the document numbers, local word numbers also begin with zero. So,
"banana": {(2, 3)} means the word "banana" is in the third document ( ), and it is the fourth word in that document (position 3).
), and it is the fourth word in that document (position 3)."a": {(2, 2)}
"banana": {(2, 3)}
"is": {(0, 1), (0, 4), (1, 1), (2, 1)}
"it": {(0, 0), (0, 3), (1, 2), (2, 0)}
"what": {(0, 2), (1, 0)}
If we run a phrase search for
"what is it" we get hits for all the words in both document 0 and 1. But the terms occur consecutively only in document 1.Applications
The inverted index data structureData structure
In computer science, a data structure is a particular way of storing and organizing data in a computer so that it can be used efficiently.Different kinds of data structures are suited to different kinds of applications, and some are highly specialized to specific tasks...
is a central component of a typical search engine indexing algorithm
Index (search engine)
Search engine indexing collects, parses, and stores data to facilitate fast and accurate information retrieval. Index design incorporates interdisciplinary concepts from linguistics, cognitive psychology, mathematics, informatics, physics, and computer science...
. A goal of a search engine implementation is to optimize the speed of the query: find the documents where word X occurs. Once a forward index is developed, which stores lists of words per document, it is next inverted to develop an inverted index. Querying the forward index would require sequential iteration through each document and to each word to verify a matching document. The time, memory, and processing resources to perform such a query are not always technically realistic. Instead of listing the words per document in the forward index, the inverted index data structure is developed which lists the documents per word.
With the inverted index created, the query can now be resolved by jumping to the word id (via random access
Random access
In computer science, random access is the ability to access an element at an arbitrary position in a sequence in equal time, independent of sequence size. The position is arbitrary in the sense that it is unpredictable, thus the use of the term "random" in "random access"...
) in the inverted index.
In pre-computer times, concordances
Concordance (publishing)
A concordance is an alphabetical list of the principal words used in a book or body of work, with their immediate contexts. Because of the time and difficulty and expense involved in creating a concordance in the pre-computer era, only works of special importance, such as the Vedas, Bible, Qur'an...
to important books were manually assembled. These were effectively inverted indexes with a small amount of accompanying commentary, that required a tremendous amount of effort to produce.
In bioinformatics, inverted indexes are very important in the sequence assembly
Sequence assembly
In bioinformatics, sequence assembly refers to aligning and merging fragments of a much longer DNA sequence in order to reconstruct the original sequence. This is needed as DNA sequencing technology cannot read whole genomes in one go, but rather reads small pieces of between 20 and 1000 bases,...
of short fragments of sequenced DNA. One way to find out where fragments came from is to search for it against a reference DNA sequence. A small number of mismatches (due to differences between the sequenced DNA and reference DNA, or due to errors) can be accounted for by dividing the fragment into smaller fragments—at least one subfragment is likely to match the reference DNA sequence. The matching requires constructing an inverted index of all substrings of a certain length from the reference DNA sequence. Since the human DNA contains more than 3 billion base pairs, and we need to store a DNA substring for every index, and a 32-bit integer for index itself, the storage requirement for such an inverted index would probably be in the tens of gigabytes, just beyond the available RAM capacity of most personal computers today.
External links
- NIST's Dictionary of Algorithms and Data Structures: inverted index
- Managing Gigabytes for Java a free full-text search engine for large document collections written in Java.
- Lucene - Apache Lucene is a full-featured text search engine library written in Java.
- Sphinx Search - Open source high-performance, full-featured text search engine library used by Craig's List and others employing an inverted index.
- Example implementations on Rosetta CodeRosetta CodeRosetta Code is a wiki-based programming chrestomathy website with solutions to various programming problems in many different programming languages. It was created in 2007 by Mike Mol. Rosetta Code includes 450 programming tasks, and covers 351 programming languages...
- Caltech Large Scale Image Search Toolbox: a Matlab toolbox implementing Inverted File Bag-of-Words image search.

