In vitro compartmentalization
Encyclopedia
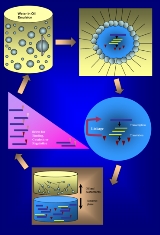
In vitro compartmentalization (IVC) is an emulsion based technology that generates cell-like compartments in vitro. These compartments are designed such that each contains no more than one gene. When the gene is transcribed and/or translated, its products (RNAs and/or proteins) become ‘trapped’ with the encoding gene inside the compartment. By coupling the genotype (DNA) and phenotype (RNA, protein), compartmentalization allows the selection and evolution of phenotype.
and phenotype
. Emulsions of cell-like compartments were formed by adding in vitro transcription
/translation
reaction mixture to stirred mineral oil containing surfactants. The mean droplet diameter was measured to be 2.6 um by laser diffraction. As a proof of concept, Tawfik el al. designed an experiment that would transcribe and translate M. HaeIII gene in the presence of 107-fold excess of genes encoding another enzyme folA. 3’ of each gene is purposely designed to contain HaeIII R/M sequences, and when HaeIII methyltransferase was expressed from a M.HaeIII gene, it would methylate HaeIII R/M sequence and cause the gene to be resistant to restriction enzyme digestion. By selecting for DNA sequences that survive the endonuclease digestion, Tawfik el al. found there was enrichment for the M.HaeIII genes, i.e. 1000 fold in the first round of selection.

(hydrophile-lipophile balance) and low HLB surfactants are needed. Some combinations of surfactants used to generate oil-surfactant mixture are mineral oil/0.5% Tween80/4.5% Span80/sodium deoxycholate and a more heat stable version, light mineral oil/0.4% Tween80/4.5% Span80/0.05%Triton-X100. The aqueous phase containing transcription and/or translation components is slowly added to the oil surfactants, and the formation of w/o is facilitated by homogenizing, stirring or using hand extruding device.
The emulsion quality can be determined by light microscopy
and/or dynamic light scattering
techniques. The emulsion is quite diverse, and greater homogenization
speeds helps to produce smaller droplets with narrower size distribution. However, homogenization speeds has to be controlled, since speed over 13,500 r.p.m tends to result in a significant loss of enzyme activity on the level of transcription. The most widely used emulsion formation gives droplets with a mean diameter of 2-3μm, and an average volume of ~5 femtoliters, or 1010 aqueous droplet per ml of emulsions. The ratio of genes to droplets is designed such that most of the droplets contains no more than a single gene statistically.
and human telomerase
.
group and an additional coding sequence for streptavidin
(STABLE display). All the newly formed proteins/peptides will be in fusion with streptavidin molecules and bind to their biotinylated coding sequence. An improved version attached two biotin molecules to the ends of a DNA molecule to increase the avidity
between DNA molecule and streptavidin-fused peptides, and used a low GC content synthetic streptavidin gene to increase efficiency and specificity during PCR amplification. The second method is to covalently link DNA and protein. Two strategies have been demonstrated. The first is to form M.HaeIII fusion proteins. Each expressed protein/polypeptide will be in fusion with Hae III DNA methyltransferase domain, which is able to bind covalently to DNA fragments containing the sequence 5’-GGC*-3’, where C* is 5-fluoro-2 deoxycytidine. The second strategy is to use monomeric mutant of VirD2 enzyme. When a protein/peptide is expressed in fusion with Agrobacterium protein VirD2, it will bind to its DNA coding sequence that has a single-stranded overhang comprising VirD2 T-border recognition sequences. The third method is to link phenotype and genotype via beads. The beads used will be coated with streptavidin to allow for the binding of biotinylated DNA, in addition, the beads will also display cognate binding partner to the affinity tag that will be expressed in fusion with the protein/peptide.
or DNA molecule. An example is the selection of proteins that have affinity to zinc finger
DNA by Sepp et al. By selecting for catalytic proteins/RNAs, new variants with novel or improved enzymatic property are usually isolated. For example, new ribozyme
variants with trans-ligase activity were selected and exhibited multiple turnovers. By selecting for regulation, inhibitors of DNA nucleases can be selected, such as protein inhibitors of the Colicin E7 DNase.
respectively.
History
In vitro compartmentalization method was first developed by Tawfik et al.. Based on the idea that Darwinian evolution relies on the linkage of genotype to phenotype, Tawfik et al. designed aqueous compartments of water-in-oil (w/o) emulsions to mimic cellular compartments that can link genotypeGenotype
The genotype is the genetic makeup of a cell, an organism, or an individual usually with reference to a specific character under consideration...
and phenotype
Phenotype
A phenotype is an organism's observable characteristics or traits: such as its morphology, development, biochemical or physiological properties, behavior, and products of behavior...
. Emulsions of cell-like compartments were formed by adding in vitro transcription
Transcription (genetics)
Transcription is the process of creating a complementary RNA copy of a sequence of DNA. Both RNA and DNA are nucleic acids, which use base pairs of nucleotides as a complementary language that can be converted back and forth from DNA to RNA by the action of the correct enzymes...
/translation
Translation
Translation is the communication of the meaning of a source-language text by means of an equivalent target-language text. Whereas interpreting undoubtedly antedates writing, translation began only after the appearance of written literature; there exist partial translations of the Sumerian Epic of...
reaction mixture to stirred mineral oil containing surfactants. The mean droplet diameter was measured to be 2.6 um by laser diffraction. As a proof of concept, Tawfik el al. designed an experiment that would transcribe and translate M. HaeIII gene in the presence of 107-fold excess of genes encoding another enzyme folA. 3’ of each gene is purposely designed to contain HaeIII R/M sequences, and when HaeIII methyltransferase was expressed from a M.HaeIII gene, it would methylate HaeIII R/M sequence and cause the gene to be resistant to restriction enzyme digestion. By selecting for DNA sequences that survive the endonuclease digestion, Tawfik el al. found there was enrichment for the M.HaeIII genes, i.e. 1000 fold in the first round of selection.
Method

Emulsion technology
Water-in-oil (w/o) emulsions are created by mixing aqueous and oil phases with the help of surfactants. A typical IVC emulsion is formed by first generating oil-surfactant mixture by stirring, and then gradually adding the aqueous phase to the oil-surfactant mixture. For stable emulsion formation, a mixture of HLBHydrophilic-lipophilic balance
The Hydrophilic-lipophilic balance of a surfactant is a measure of the degree to which it is hydrophilic or lipophilic, determined by calculating values for the different regions of the molecule, as described by Griffin in 1949 and 1954...
(hydrophile-lipophile balance) and low HLB surfactants are needed. Some combinations of surfactants used to generate oil-surfactant mixture are mineral oil/0.5% Tween80/4.5% Span80/sodium deoxycholate and a more heat stable version, light mineral oil/0.4% Tween80/4.5% Span80/0.05%Triton-X100. The aqueous phase containing transcription and/or translation components is slowly added to the oil surfactants, and the formation of w/o is facilitated by homogenizing, stirring or using hand extruding device.
The emulsion quality can be determined by light microscopy
Microscopy
Microscopy is the technical field of using microscopes to view samples and objects that cannot be seen with the unaided eye...
and/or dynamic light scattering
Dynamic light scattering
thumb|right|350px|Hypothetical Dynamic light scattering of two samples: Larger particles on the top and smaller particle on the bottomDynamic light scattering is a technique in physics that can be used to determine the size distribution profile of small particles in suspension or polymers...
techniques. The emulsion is quite diverse, and greater homogenization
Homogenization (chemistry)
Homogenization or homogenisation is any of several processes used to make a chemical mixture the same throughout.-Definition:Homogenization is intensive blending of mutually related substances or groups of mutually related substances to form a constant of different insoluble phases to obtain a...
speeds helps to produce smaller droplets with narrower size distribution. However, homogenization speeds has to be controlled, since speed over 13,500 r.p.m tends to result in a significant loss of enzyme activity on the level of transcription. The most widely used emulsion formation gives droplets with a mean diameter of 2-3μm, and an average volume of ~5 femtoliters, or 1010 aqueous droplet per ml of emulsions. The ratio of genes to droplets is designed such that most of the droplets contains no more than a single gene statistically.
In Vitro transcription/translation
IVC has used bacterial cell, wheat germ and rabbit reticulocyte (RRL) extracts for transcription and translation. It is also possible to use bacterial reconstituted translation system such as PURE in which translation components are individually purified and later combined. When expressing eukaryote or complex proteins, it is desirable to use eukaryotic translation systems such as wheat germ extract or more superior alternative, RRL extract. In order to use RRL for transcription and translation, traditional emulsion formulation cannot be used as it abolishes translation. Instead, a novel emulsion formulation: 4% Abil EM90 / light mineral oil was developed and demonstrated to be functional in expressing luciferaseLuciferase
Luciferase is a generic term for the class of oxidative enzymes used in bioluminescence and is distinct from a photoprotein. One famous example is the firefly luciferase from the firefly Photinus pyralis. "Firefly luciferase" as a laboratory reagent usually refers to P...
and human telomerase
Telomerase
Telomerase is an enzyme that adds DNA sequence repeats to the 3' end of DNA strands in the telomere regions, which are found at the ends of eukaryotic chromosomes. This region of repeated nucleotide called telomeres contains non-coding DNA material and prevents constant loss of important DNA from...
.
Breaking emulsion and coupling of genotype and phenotype
Once transcription and/or translation has completed in the droplets, emulsion will be broken by successive steps of removing mineral oil and surfactants to allow for subsequent selection. At this stage, it is crucial to have a method to ‘track’ each gene products to the encoding gene as they become free floating in a heterogeneous population of molecules. There are three major approaches to track down each phenotype to its genotype. The first method is to attach each DNA molecule with a biotinBiotin
Biotin, also known as Vitamin H or Coenzyme R, is a water-soluble B-complex vitamin discovered by Bateman in 1916. It is composed of a ureido ring fused with a tetrahydrothiophene ring. A valeric acid substituent is attached to one of the carbon atoms of the tetrahydrothiophene ring...
group and an additional coding sequence for streptavidin
Streptavidin
Streptavidin is a 60000 dalton protein purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin . With a dissociation constant on the order of ≈10-14 mol/L, the binding of biotin to streptavidin is one of the strongest...
(STABLE display). All the newly formed proteins/peptides will be in fusion with streptavidin molecules and bind to their biotinylated coding sequence. An improved version attached two biotin molecules to the ends of a DNA molecule to increase the avidity
Avidity
In proteins, avidity is a term used to describe the combined strength of multiple bond interactions. Avidity is distinct from affinity, which is a term used to describe the strength of a single bond...
between DNA molecule and streptavidin-fused peptides, and used a low GC content synthetic streptavidin gene to increase efficiency and specificity during PCR amplification. The second method is to covalently link DNA and protein. Two strategies have been demonstrated. The first is to form M.HaeIII fusion proteins. Each expressed protein/polypeptide will be in fusion with Hae III DNA methyltransferase domain, which is able to bind covalently to DNA fragments containing the sequence 5’-GGC*-3’, where C* is 5-fluoro-2 deoxycytidine. The second strategy is to use monomeric mutant of VirD2 enzyme. When a protein/peptide is expressed in fusion with Agrobacterium protein VirD2, it will bind to its DNA coding sequence that has a single-stranded overhang comprising VirD2 T-border recognition sequences. The third method is to link phenotype and genotype via beads. The beads used will be coated with streptavidin to allow for the binding of biotinylated DNA, in addition, the beads will also display cognate binding partner to the affinity tag that will be expressed in fusion with the protein/peptide.
Selection
Depending on the phenotype to be selected, difference selection strategies will be used. Selection strategy can be divided into three major categories: selection for binding, selection for catalysis and selection for regulation. The phenotype to be selected can range from RNA to peptide to protein. By selecting for binding, the most commonly evolved phenotypes are peptide/proteins that have selective affinity to a specific antibodyAntibody
An antibody, also known as an immunoglobulin, is a large Y-shaped protein used by the immune system to identify and neutralize foreign objects such as bacteria and viruses. The antibody recognizes a unique part of the foreign target, termed an antigen...
or DNA molecule. An example is the selection of proteins that have affinity to zinc finger
Zinc finger
Zinc fingers are small protein structural motifs that can coordinate one or more zinc ions to help stabilize their folds. They can be classified into several different structural families and typically function as interaction modules that bind DNA, RNA, proteins, or small molecules...
DNA by Sepp et al. By selecting for catalytic proteins/RNAs, new variants with novel or improved enzymatic property are usually isolated. For example, new ribozyme
Ribozyme
A ribozyme is an RNA molecule with a well defined tertiary structure that enables it to catalyze a chemical reaction. Ribozyme means ribonucleic acid enzyme. It may also be called an RNA enzyme or catalytic RNA. Many natural ribozymes catalyze either the hydrolysis of one of their own...
variants with trans-ligase activity were selected and exhibited multiple turnovers. By selecting for regulation, inhibitors of DNA nucleases can be selected, such as protein inhibitors of the Colicin E7 DNase.
Advantages
Comparing to other in vitro display technologies, IVC has two major advantages. The first advantage is its ability to control reactions within the droplets. Hydrophobic and hydrophilic components can be delivered to each droplet in a step-wise fashion without compromising the chemical integrity of the droplet, and thus by controlling what to be added and when to be added, the reaction in each droplet is controlled. In addition, depending on the nature of the reaction to be carried out, the pH of each droplet can also be changed. More recently, photocaged substrates were used and their participation in a reaction was regulated by photo-activation. The second advantage is that IVC allows the selection of catalytic molecules. As an example, Griffiths et al. was able to select for phosphotriesterase variants with higher Kcat by detecting product formation and amount using anti-product antibody and flow cytometryFlow cytometry
Flow cytometry is a technique for counting and examining microscopic particles, such as cells and chromosomes, by suspending them in a stream of fluid and passing them by an electronic detection apparatus. It allows simultaneous multiparametric analysis of the physical and/or chemical...
respectively.
Related technologies
- CIS display
- Phage displayPhage displayPhage display is a method for the study of protein–protein, protein–peptide, and protein–DNA interactions that uses bacteriophages to connect proteins with the genetic information that encodes them. Phage Display was originally invented by George P...
- Bacterial displayBacterial displayBacterial display is a protein engineering technique used for in vitro protein evolution...
- Yeast displayYeast displayYeast display is a technique used in the field of protein engineering. The yeast display technique was first published by the laboratory of Professor K. Dane Wittrup. The technology was sold to Abbott Laboratories in 2001....
- Ribosome displayRibosome displayRibosome display is a technique used to perform in vitro protein evolution to create proteins that can bind to a desired ligand. The process results in translated proteins that are associated with their mRNA progenitor which is used, as a complex, to bind to an immobilized ligand in a selection step...
- mRNA displayMRNA displaymRNA display is a display technique used for in vitro protein, and/or peptide evolution to create molecules that can bind to a desired target. The process results in translated peptides or proteins that are associated with their mRNA progenitor via a puromycin linkage. The complex then binds to...

